1. What is taxonomy, what’s the reality behind it – and why do we need it anyway?
Taxonomy is the branch of science concerned with the classification of organisms. Since Darwin, it has allowed to understand the relationship between species in the form of the Tree of Life. However, categorizing is an exquisitely human activity and reflects more the functioning of the human mind than the reality of Life itself. A quote of Stephen Jay Gould clearly portrays the relevance of taxonomy for human thinking as “the way we order reflects the way we think. Historical changes in classification are the fossilized indicators of conceptual revolutions” (Gould SJ. 1983).
Therefore, the anticipated reclassification of the Genus Lactobacillus – apart from the technical and legal difficulties, harbors the potential for a conceptual revolution within the academic and industrial fields.
The Tree of Life described by Darwin in On The Origin Of Species can be seen as a branching structure of life resulting from descent with modification. When looking at animals and plants, this hypothesis has proven quite nailed down. However, when it comes down to prokaryotes (isolated cells without a nucleus, like Bacteria and Archea) we are confronted with such small and invisible entities that a different approach is needed. Because bacteria don’t just divide to create a second generation (vertical transmission of the genes) but also continuously exchange genetic material through conjugations with other bacteria in their ecosystem (horizontal transmission) – and especially in the human intestinal microbiome (Jeong H, et al., 2019), their Tree of Life is much more intertwined. Darwin’s model of evolution is not able to accommodate for the existence and abundance of this Horizontal Gene Transfer (HGT) – that threatens the very concept of a Bacterial Tree of Life.
Indeed, as reported by Sorin Sonea in the foreword of the highly pioneering 1983 “A new bacteriology”: bacteria “function in the world by acquiring and exchanging a diverse set of variable genetic systems.” A modern view of the prokaryote world is indeed conceptualized as a tangled “Web of life” more than a Treeeven though the Tree is adopted as a convenient determined structure for classification (and thus visualization) purposes (Soucy SM, et al., 2015).
In fact, any attempt of taxonomical classification implicitly relies on a specific microorganism which has been isolated from its ecosystem to the lab and “frozen in time”, thus limiting the natural occurrence of HGT within the microbial community. In other words, the Human activity of observing the microorganisms unavoidably decontextualizes its natural genomic fluidity and this defines the boundaries of we call a species or even a genus in the bacterial world.
Notably, adopting new metrics also reveals previously unseen organismal relationships, highlighting the dynamic state of the prokaryotic taxonomy.Of course, the truth of our models is related to the methodologies used to decode the reality. Only with the advent of molecular techniques did scientists have the possibility to draw bacterial phylogenetic trees.
However, despite its arbitrary component, taxonomy is really necessary for Human activities, as its core objective is to give a common (and preferably long-lasting) name to an object (both concrete or abstract) which can be shared across the community.
2. How do scientists classify microorganisms nowadays and what’s the history behind this branch of taxonomy?
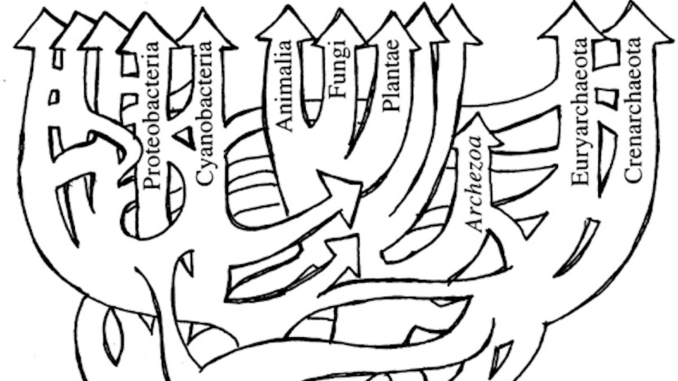
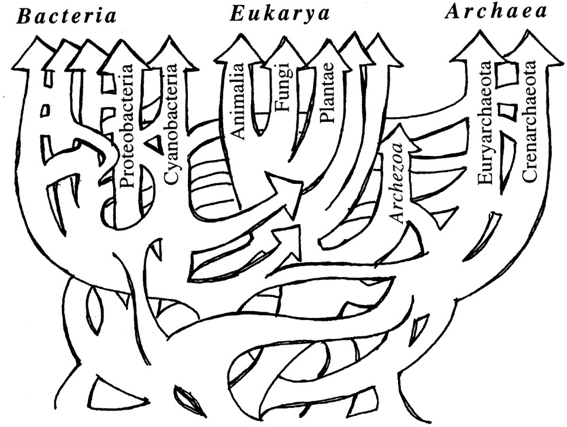
Bacterial nomenclature has been a continuous challenge since the first observation by Antoni van Leeuwenhoek in 1676 of small “animalcules” under his home-made microscope. The small dimension, the inability to grow in pure culture for most of the microorganisms – 99.5% are not cultivable! (Harwani D. 2012; Lloyd KG, et al., 2018) -, the absence of sexual reproduction and their peculiar capability to share horizontally genetic elements have generated frustration between taxonomists who tried to define a conceptually elusive Bacterial Kingdom which is and acts more properly as a web-of-life (Fig.1) rather than being composed by definite members exhibiting fixed, distinctive traits (Swithers KS,et al., 2009).
In the end of the 19thcentury, Pasteur was able to prove the bacterial origin of several infectious diseases in animals which led to the development of methods for discovery, isolation and characterization of the majority of disease-causing bacteria. These bacteria were called pathogens from pathos (illness) and gen (causing). Bacteria were then seen mostly as a treat and therefore most of the research was directed towards the prevention or treatment of bacterial infection, as well as diagnosis and immunization. It was thanks to this effort that a large portion of humanity was freed from deadly infectious disease, permitting an increase of human life expectancy and generating the population explosion. Interestingly the practical success in the fight against bacterial disease generated a lasting decrease in scientific interest in bacteria per se(something which is trending back currently thanks to microbiome studies). The research focus on pathogens meant that identifying the isolate, as linked to Koch’s postulate, was crucial to treat. This characterization in the clinical and laboratory setting, however, led to fixism: the theory that bacterial strains do not change in nature.Consequently, the concept of fixed bacterial species became the paradigm.
From the mid-19thcentury to the 1970s bacterial characterization and classification relied mostly on phenotypic, morphological and chemotaxonomic traits. Then Carl Woese, bombarding Bacillus subtilis spores with ionizing radiation in the 1950s and looking ever more deeply into ribosome particles and RNA properties of germinating spores, made the discovery with George Fox of the three domains of Life – Bacteria, Archaea and Eukarya (Woese C and Fox G, 1977). It is impressive to think that Woese, deemed the father of 16s rRNA-based phylogenetics, used rRNA as an evolutionary marker nearly 10 years before a technique for efficient 16s gene sequencing was developed! (Zhulin IB, 2016).
Over time, evolutionary researchers have proposed different models of trees; first at the scale of genes (16s RNA, a gene coding for a subpart of the ribosome that translates DNA into RNA, is the most famous gene used for this kind of classification because is it universally distributed and sufficiently conserved) and more recently at the scale of genomes (thanks to the technical advancement and availability of complete genome sequences). We cannot imagine current bacterial taxonomy without phylogenies derived from comparison of small-subunit 16S rRNA gene sequences the use of a single gene (eg. 16s rRNA) may not reflect the evolution history of the genome. In fact, 16s rRNA constitutes approximately 0.07% of the whole genome, and a mere 1,3% of difference within the 16rRNA sequence corresponds to 50 million years of evolutionary divergence! (Ochman H,et al., 1999). Using a different marker gene could lead to a different construction of relatedness. Therefore, evolutionary relatedness, inferred from phylogenetic markers like 16s rRNA, facilitates classification but does not represent the whole picture. With both sequencing and analysis techniques rapidly improving, microbial taxonomy may be in the beginning of its third phase – phylogenomics – which relies on genome-wide comparisons of genes (Kislyuk AO, et al., 2011) and/or proteins (Callister SJ, et al., 2008).
Moreover, there are non-phylogenetic alternatives for classification. Phenetics, for example, classifies organisms based on functional similarity regardless of shared ancestry. It is important to note that though both phylogeny-based taxonomy and phenetics can be used to investigate bacterial relationships, the questions that they try to answer are different. The task of phylogeny is reconstructing organismal evolutionary history efforts while Phenetics clusters organisms into currently consistent classes on the basis of observable traits. Closely related organisms are often phenotypically similar, but not always (Zhu C, et al., 2015; Sterner B, et al., 2018).
Through history, the continuous technological and methodological development has given to scientist’s ever more modern tools to decipher and classify prokaryotes, which inevitably have brought numerous times to the redefinition of bacterial taxonomy. In 1966 for example, a bacterial census enumerated 28 900 different species. Better methods and criteria allowed to reduce these numbers down to 1792 just 14 years later (Skerman VBD, et al., 1980). In 2017, with the identification of many new microorganisms, this number had risen up again to 15,626 species, listed in the official reference List of Prokaryotic names with Standing in Nomenclature (LPSN), available on www.bacterio.net.Bacterial Taxonomy is regulated by the International Code of Nomenclature of Prokaryotes which includes 65 rules designated to assess the correctness of a bacterial name (Parker CT, et al., 2015) and is recognized internationally.
3. What about Lactobacilli? Why change their classification, and on which grounds?
While reclassification happened before, it was mostly a matter of scientific discussion, isolated within the taxonomists room, or a matter of safety concern linked to the identification of pathogenic bacteria. Now and for the first time, the upcoming taxonomy change will have an unprecedented impact on probiotics – a category consumed by millions of people Worldwide and projected to reach almost 70 billion USD of market value by 2023 (Markets and Markets).
Lactobacilli are widely administered in food and food supplements to healthy and less healthy populations, with implications that encompass safety, efficacy, link to scientific publications, intellectual property and of course commercial interests.
Despite the existence of a major impacted market, the tremendous genomic diversity between the over 200 species belonging to the genus Lactobacillus and the continuous addition of novel species calls the taxonomists to review the current classification (Fig.2).
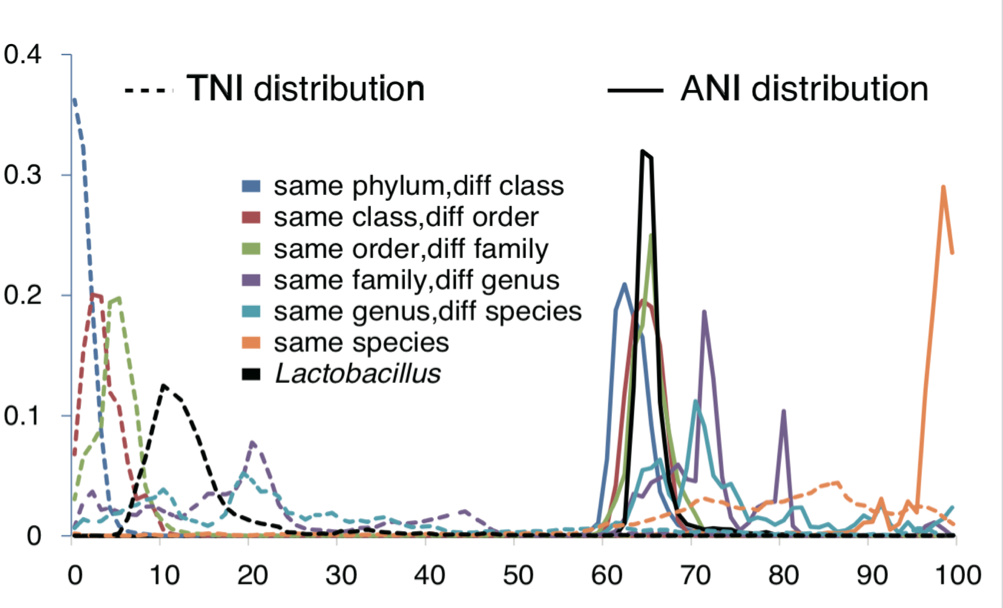
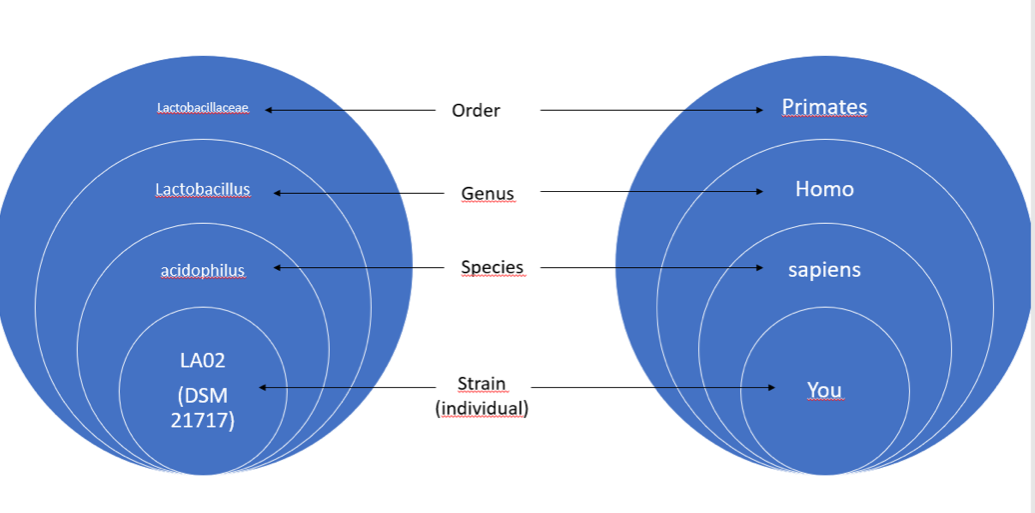
This wide genetic diversity means that we currently group together under the same genus bacteria that are as different as a man is from a lemur (Fig. 3).
With the use of different methodologies (distance-based metrics; average nucleotide identity; average amino-acid identity; percentage of conserved proteins; phylogenetic and network analysis on proteins and phylogenetic markers on 222 different Lactobacillus strains), Salvetti E. et al., 2018 presented a robust proposal to organize the genus Lactobacillus in 10 consistent subgroups, namely, (i) L. delbrueckii (named after the type species of Lactobacillus), which also comprises the peripheral species Lactobacillus amylophilus, Lactobacillus amylotrophicus, and Lactobacillus floricola; (ii) L. alimentarius; (iii) L. perolens: (iv) L. casei; (v) L. sakei (without L. selangorensis); (vi) L. coryniformis; (vii) L. salivarius; (viii) L. plantarum; (ix) L. reuteri, which also includes L. vaccinostercus-related species; and (x) L. buchneri, which encompasses members of the L. brevis, L. fructivorans, and L. collinoides groups.
Another proposal from Parks DH, et al. also in 2018, was based on a phylogenetic analysis of 120 proteins on the staggering number of 94,759 prokaryotic genomes, and generated 16 different groups within the Lactobacillus genus (https://gtdb.ecogenomic.org).
Previously, Zheng J, et al.had clustered in 2015 24 new subgroups among 174 Lactobacillus and Pediococcus type strains based on 172 single-copy core genes involved in ecology and biochemistry properties.
In the end No True Taxonomy Exists and depending on the methodology and cut-off used, different groups can be generated – for example 10 based on Salvetti’s work, 16 in Parks and 24 in Zhengs!
The two key aspects for the choice of the new classification will be to create new genera with a better homogeneity that respects the defined rules for similarity between organisms in a same genus, and most importantly to create a stable number of genera that will accommodate future species without the need of a further reclassification.
Moreover, these future smaller groups could help to focus easier on shared functional characteristics based on physiological properties, biogeography, ecology, natural history and potential interaction with the hosts in order to fill the gap on molecular mechanisms of action and ecologic fitness – which can help understand better the bacteria and their use.
4) What are the benefits behind the taxonomy change?
The Lactobacillus reclassification will be a hurdle that will mean for many stakeholders significant investments for the update of labels, quality and technical documentation, certificates and export approvals, agreements, Intellectual Property, marketing tools, websites, etc. It will be a challenge to communicate to customers and consumers that the names of bacteria on the products is changing though the bacteria themselves aren’t, but it will also be a unique occasion to better explain the world of bacteria, to better understand the specificities among the new groups, and to better develop targeted products with a deeper functional vision and ultimately, better efficacy.
The evolutionary history of bacteria determines how they interact with the host, and for this reason is relevant for the selection of Lactobacilli for therapeutic applications. For example, L. plantarum, L. casei and L. fermentum are nomadic and free-living by history, while L. reuteri, L. johnsonii and L. acidophilus are host-adapted species (Duar RM, et al., 2017). That implies that the nomadic bacteria have a wider genome and more tools to survive in various environments. Host-adapted species are more competitive when compared to bacteria that do not share an evolutionary history with the host.
This is a strength if a probiotic is selected to antagonize a pathogen. On the contrary, if the aim is to stimulate the immune system, the selection of a probiotic without a joint evolution with the host may be a more interesting approach, as it will be potentially less tolerated and more inclined to stimulate an immune response.
Adaptation to a host often means a shedding of genes that are no longer needed in the symbiotic lifestyle. Interestingly, among Lactobacilli the species with the highest degree of host specialization are the ones occurring in the human vagina: L. iners, L. crispatus, L. jensenii and L. gasseri, with L. iners and jensenii only found in this niche. It is likely this has an effect on possible survival through oral intake, due to the loss of capacity to survive gastric acidity or to adhere in the gut epithelium for example, and this questions the regulatory limitations in Europe towards medical devices, since vaginally applied probiotics will no longer be authorized for commercialization as of May 2020. From the above example, even if the substance (the microorganisms) is the same, adopting a new higher taxonomical name will help to demarcate conceptually its new biogeography, projecting an easier experimental hypothesis on the functional role of the strain of interest to be verified.
More largely and futuristically, a better understanding of the bacteria can allow the development of strategies to support their populations or even specific activities through dietary intervention.
All in all, what can you expect?
Uniting the clues collected from the last taxonomy articles, we can hypothesize that the Lactobacillus genus will be split into at least 20 new genera. Keep an eye on the website http://bacterio.net/index.html for the definitive answer! The new classification will be a more precise vision of clusters of bacteria sharing common characteristics, resulting from a consensus between taxonomists, itself allowed by technological advances. It will aim to ensure a shared and stable language to identify bacteria correctly, and will be a historic event leading for the first time in the field of taxonomy to a global impact on a pre-existing economy of over 50 billion dollars. As any change of categorization translates a change of paradigm, there will be new perspectives leading to new ways of developing probiotic products.
By the way, next in line, Bifidobacteria await for their new look too!
Bibliography:
Callister SJ, McCue LA, Turse JE, Monroe ME, Auberry KJ, Smith RD et al. (2008). Comparative bacterial proteomics: analysis of the core genome concept. PLoS One 3: e1542.
Ciufo S, Kannan S, Sharma S, Badretdin A, Clark K, Turner S, Brover S, Schoch CL, Kimchi A, DiCuccio M. Using average nucleotide identity to improve taxonomic assignments in prokaryotic genomes at the NCBI. Int J Syst Evol Microbiol. 2018 Jul;68(7):2386-2392. doi: 10.1099/ijsem.0.002809. Epub 2018 May 24. PubMed PMID: 29792589.
Doolittle WF. Phylogenetic classification and the universal tree. Science. 1999 Jun 25;284(5423):2124-9. Review. PubMed PMID: 10381871.
Duar RM, Lin XB, Zheng J, Martino ME, Grenier T, Pérez-Muñoz ME, Leulier F, Gänzle M, Walter J. Lifestyles in transition: evolution and natural history of the genus Lactobacillus. FEMS Microbiol Rev. 2017 Aug 1;41(Supp_1):S27-S48. doi:10.1093/femsre/fux030. Review. PubMed PMID: 28673043.
Gould SJ. 1983. Hen’s teeth and horse’s toes. Norton, New York, NY.
Harwani, Dharmesh. (2012). The Great Plate Count Anomaly and the Unculturable Bacteria. International Journal of Scientific Research. 2. 350-351. 10.15373/22778179/SEP2013/122.
ISO 20128:2006 IDF 192:2006 https://www.iso.org/standard/35292.html
ISO 7889:2003 IDF 117:2003 https://www.iso.org/standard/31880.html
ISO 19344:2015 IDF 232:2015 https://www.iso.org/standard/64658.html
Jeong H, Arif B, Caetano-Anollés G, Kim KM, Nasir A. Horizontal gene transfer in human-associated microorganisms inferred by phylogenetic reconstruction and reconciliation. Sci Rep. 2019 Apr 11;9(1):5953. doi: 10.1038/s41598-019-42227-5. PubMed PMID: 30976019; PubMed Central PMCID: PMC6459891.
Kim M, Oh HS, Park SC, Chun J. Towards a taxonomic coherence between average nucleotide identity and 16S rRNA gene sequence similarity for species demarcation of prokaryotes. Int J Syst Evol Microbiol. 2014 Feb;64(Pt 2):346-51. doi: 10.1099/ijs.0.059774-0. Erratum in: Int J Syst Evol Microbiol. 2014 May;64(Pt 5):1825. PubMed PMID: 24505072.
Kislyuk AO, Haegeman B, Bergman NH, Weitz JS (2011). Genomic fluidity: an integrative view of gene diversity within microbial populations. BMC Genomics 12: 32.]
Lloyd KG, Steen AD, Ladau J, Yin J, Crosby L. Phylogenetically Novel Uncultured Microbial Cells Dominate Earth Microbiomes. mSystems. 2018 Sep 25;3(5). pii: e00055-18. doi: 10.1128/mSystems.00055-18. eCollection 2018 Sep-Oct. PubMed PMID: 30273414; PubMed Central PMCID: PMC6156271.
Markets and Markets: https://www.marketsandmarkets.com/Market-Reports/probiotic-market-advanced-technologies-and-global -market-69.html).
Ochman H, Elwyn S, Moran NA. Calibrating bacterial evolution. Proc Natl AcadSci U S A. 1999 Oct 26;96(22):12638-43. PubMed PMID: 10535975; PubMed Central PMCID: PMC23026.
Parker CT, Tindall BJ, Garrity GM. 2015. International Code of Nomenclature of Prokaryotes. Int J Syst Evol Microbiol 68:1825–1829. https:// doi.org/10.1099/ijsem.0.000778.
Parks DH, Chuvochina M, Waite DW, Rinke C, Skarshewski A, Chaumeil PA, Hugenholtz P. A standardized bacterial taxonomy based on genome phylogeny substantially revises the tree of life. Nat Biotechnol. 2018 Nov;36(10):996-1004. doi: 10.1038/nbt.4229. Epub 2018 Aug 27. PubMed PMID: 30148503.
Salvetti E, Harris HMB, Felis GE, O’Toole PW. Comparative Genomics of the Genus Lactobacillus Reveals Robust Phylogroups That Provide the Basis for Reclassification. Appl Environ Microbiol. 2018 Aug 17;84(17). pii: e00993-18. doi: 10.1128/AEM.00993-18. Print 2018 Sep 1. Erratum in: Appl Environ Microbiol. 2018 Oct 1;84(20):. PubMed PMID: 29915113; PubMed Central PMCID: PMC6102987.
Skerman VBD, McGowan V, Sneath PHA. Approved lists of bacterial names. Int J Syst Evol Microbiol. 1980;30:225–420.
Sorin Sonea and Maurice Panisset, A New Bacteriology, 1983, Published by Jones and Bartlett Learning.
Soucy SM, Huang J, Gogarten JP. Horizontal gene transfer: building the web of life. Nat Rev Genet. 2015 Aug;16(8):472-82. doi: 10.1038/nrg3962. Review. PubMed PMID: 26184597.
Sterner B, Lidgard S. Moving Past the Systematics Wars. J Hist Biol. 2018 Mar;51(1):31-67. doi: 10.1007/s10739-017-9471-1. PubMed PMID: 28255641.
Sun Z, Harris HM, McCann A, Guo C, Argimón S, Zhang W, Yang X, Jeffery IB, Cooney JC, Kagawa TF, Liu W, Song Y, Salvetti E, Wrobel A, Rasinkangas P, Parkhill J, Rea MC, O’Sullivan O, Ritari J, Douillard FP, Paul Ross R, Yang R,Briner AE, Felis GE, de Vos WM, Barrangou R, Klaenhammer TR, Caufield PW, Cui Y, Zhang H, O’Toole PW. Expanding the biotechnology potential of lactobacilli through comparative genomics of 213 strains and associated genera. Nat Commun. 2015 Sep 29;6:8322. doi: 10.1038/ncomms9322. PubMed PMID: 26415554; PubMedCentral PMCID: PMC4667430.]
Swithers KS, Gogarten JP, Fournier GP. Trees in the web of life. J Biol. 2009;8(6):54. doi: 10.1186/jbiol160. Epub 2009 Jul 13. Review. PubMed PMID: 19664165; PubMed Central PMCID: PMC2737374.
Woese CR, Fox GE.1977. Phylogenetic structure of the prokaryoticdomain: the primary kingdoms. Proc Natl Acad SciUSA74:5088 –5090.http://dx.doi.org/10.1073/pnas.74.11.5088.
Zheng J, Ruan L, Sun M, Gänzle M. A Genomic View of Lactobacilli and Pediococci Demonstrates that Phylogeny Matches Ecology and Physiology. Appl Environ Microbiol. 2015 Oct;81(20):7233-43. doi: 10.1128/AEM.02116-15. Epub 2015 Aug 7. PubMed PMID: 26253671; PubMed Central PMCID: PMC4579461.
Zhu C, Delmont TO, Vogel TM, Bromberg Y. Functional Basis of Microorganism Classification. PLoS Comput Biol. 2015 Aug 28;11(8):e1004472. doi: 10.1371/journal.pcbi.1004472. eCollection 2015 Aug. PubMed PMID: 26317871; PubMed Central PMCID: PMC4552647.
Zhulin IB. 2016. Classic spotlight: 16S rRNA redefines microbiology.J Bacteriol 198:2764-2765. doi:10.1128/JB.00616-16