Microbiome research promises to transform human health; however, until now the field has been held back by the challenges of translating microbiome data into novel treatments, ingredients, diagnostics, and other healthcare solutions.
A number of high-impact microbiome studies have been criticized for fundamental flaws in their profiling results. In one case, as alleged by Gihawi et al. (2023), numerous artificial detections of bacteria in tumors – notably, bacteria that are not normally associated with humans – were reported. The inaccurate profiling invalidated not only the results of the original study but also a dozen additional published studies building on the same erroneous data, highlighting the importance of correct microbiome profiling.
Clinical Microbiomics introduces its human microbiome profiler 2.0: CHAMP™, a best-in-class tool for human microbiome profiling. Developed by Dr. H. Bjørn Nielsen and his team of bioinformatics experts, CHAMP™ sets new standards in sensitivity and accuracy for microbiome analysis.
“With CHAMP™️, we introduce our next-generation human microbiome profiler. It’s based on the world’s largest collection of high-quality human microbiome data, encompasses the full microbial diversity across all body sites, and assures profiling with unmatched accuracy and incredibly low levels of false signals.”
Dr. H. Bjørn Nielsen comments.
Microbiome profiling pipelines should analyse microbiome composition with emphasis on the following:
|
De-risking microbiome research and clinical development
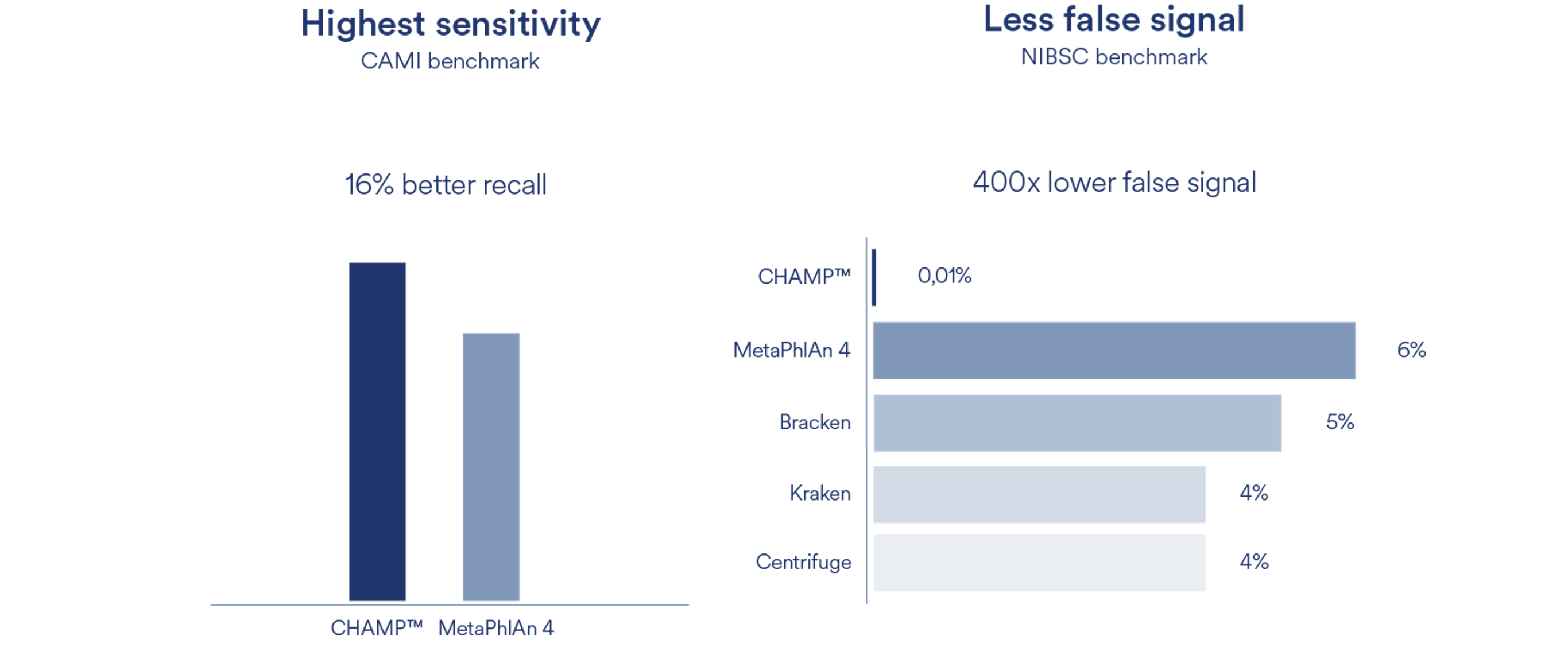
CHAMP™ provides high-resolution taxonomic and functional annotation from shotgun sequencing data and offers the highest specificity, sensitivity and most accurate profiling when benchmarked against the best and most widely used profiling pipelines in the field.
Compared to MetaPhlAn4, CHAMP™ showed 16% greater sensitivity (recall) across different human body sites and showed an astounding 400 times lower false signal in the NIBSC mock community benchmark compared to state-of-the-art profilers (MetaPhlAn4, Centrifuge, Kraken, and Bracken). This means that when CHAMP™ detects something, you can trust it’s there. In contrast, Kraken detects almost 100 species in a mock community that contains only 20. Such false signals are detrimental in the development of new probiotics and microbiome-based therapeutics, with incorrect bacterial candidates brought forward into costly clinical trials, and their mechanisms of action misunderstood.
Dr. H. Bjørn Nielsen emphasizes:
“Correct profiling is the cornerstone of a sound microbiome study – without it, conclusions can easily be misguided.”
Clinical Microbiomics’ extensive reference database and intelligent profiling algorithm makes CHAMP™ the best starting-point for human microbiome research
CHAMP™ was built using Clinical Microbiomics’ extensive library of human microbiome data, the most comprehensive reference catalogue in the world for human-associated microbes. Clinical Microbiomics’ database comprises over 400,000 Metagenome Assembled Genomes (MAGs) collected across 9 body sites including but not limited to: stool, small intestine, the vagina, the skin and the mouth. To ensure results are robust to the natural strain diversity across human microbiome samples, CHAMP™ leverages a proprietary algorithm calibrated across thousands of analyzed samples.
CHAMP™ can be deployed across a range of use-cases to ensure accurate and sensitive microbiome analysis.
The development of microbiome therapeutics is likely to require validated microbiome profiling. To be validated for clinical trials, a drug developer’s profiling pipeline must be demonstrably consistent in its performance. CHAMP™ builds upon previous versions of Clinical Microbiomics’ microbiome profilers that have been used in several phase 2 and 3 clinical trials from high-profile drug developers, demonstrating its quality and reliability in the eyes of regulators.
For skin and vaginal microbiome research, the low-biomass, high-host nature of samples results in few microbial reads from shotgun sequencing. Therefore, to obtain an accurate understanding of the microbiome of these body sites, a highly sensitive and specific profiling pipeline is necessary. With its superior specificity and sensitivity CHAMP™ offers researchers confidence that all species, even low abundant ones, will be detected.
CHAMP™ also uses the latest GTDB annotation, which includes more rare species and compared to NCBI often classify sub-species as species in their own right. This level of resolution is key to understanding the mechanisms at play, its importance exemplified in the study of the infant microbiome. Here, multiple similar species and subspecies of Bifidobacterium have unique carbohydrate-utilization patterns which are crucial for the infants’ health, but this diversity is missed by common microbiome profilers.
The future of microbiome research is here
CHAMP™ is a revolutionary microbiome profiling tool that excels in high sensitivity, high accuracy, and low false signal. Benchmarking results show superior performance compared to current state-of-the-art profiling technologies. With its extensive reference database and proprietary algorithm, CHAMP™ is made for use across human microbiome research, including in clinical trials and drug development. What’s more, Clinical Microbiomics are continuing to build out the capabilities of their profiling platform with expanded phage profiling and advanced functional annotation currently in development.
To find out more about CHAMP™ click here.

