
The list of presentations from any recent microbiome conference shows the diversity of approaches by which scientists are pursuing discovery of microbiome-focused therapeutics. And common to all of these approaches is the essential need to predict how microbial communities might work, in all their complexity, to influence host health.
Various mathematical models are used to capture aspects of complex microbial ecosystems and hosts. But which aspects? The give-and-take between hosts and microbial communities over time in response to a changing environment may be important for determining certain health-related phenotypes. So some scientists are arguing evolution of the holobiont – a host and its microbes considered as one unit – may be important to model for a better understanding of health – or more specifically, of how to manipulate the microbiome for a particular therapeutic outcome.
“…understanding the process of the emergence of symbiosis and co-dependence is essential in understanding human evolution and health.”
As humans have evolved, microbes have always been their ‘guests’. And as microbial hosts, humans have developed microbe-controlling mechanisms (like the immune system) and have selected microbes that perform certain jobs for them. Humans and microbes thus depend on each other for survival – so can we gain a better understanding of how this symbiosis arose in the first place, and what might trigger a shift to a dysbiotic state?
Dr. Alejandro Frank (AF), a scientist at Centro de Ciencias de la Complejidad, Universidad Nacional Autónoma de México, in Mexico City (Mexico) recently addressed similar questions in a paper in Frontiers in Physiology, co-authored with Mexican colleagues, that proposed a mathematical model of the general mechanisms of holobiont evolution. In this interview, Frank tells Microbiome Times about his group’s approach and how models like theirs could be used to increase knowledge of both symbiosis and dysbiosis and of microbe-host interactions for health.
What unique perspectives can be gained by considering holobiont evolution?
AF: The process of evolution in the holobiont and the forces that drive it are (and should be) the same as in any other evolutionary processes. For instance, genetic mutation, genetic drift, and gene flow are clearly present, and influenced by natural selection.
However, these forces may apply at different timescales and rates for different parts of the holobiont, as the bacterial, viral and eukaryotic parts do not work in the same way. We believe that using this more complex, ecological approach, is what makes the holobiont a unique perspective.
With your recent paper, which aspect of holobiont evolution were you hoping to address?
AF: Our intention was to elucidate why and how symbiosis appears by using a relatively simple model of the process of mutation and coexistence between microbial communities and their hosts.
It was also our goal to understand why the microbial communities help their hosts and why they tend to lose independence on the way, as many symbiotic microbes cannot live without their hosts. Host and microbes develop a co-dependence. We believe that understanding the process of the emergence of symbiosis and co-dependence is essential in understanding human evolution and health.
How did you model this mathematically?
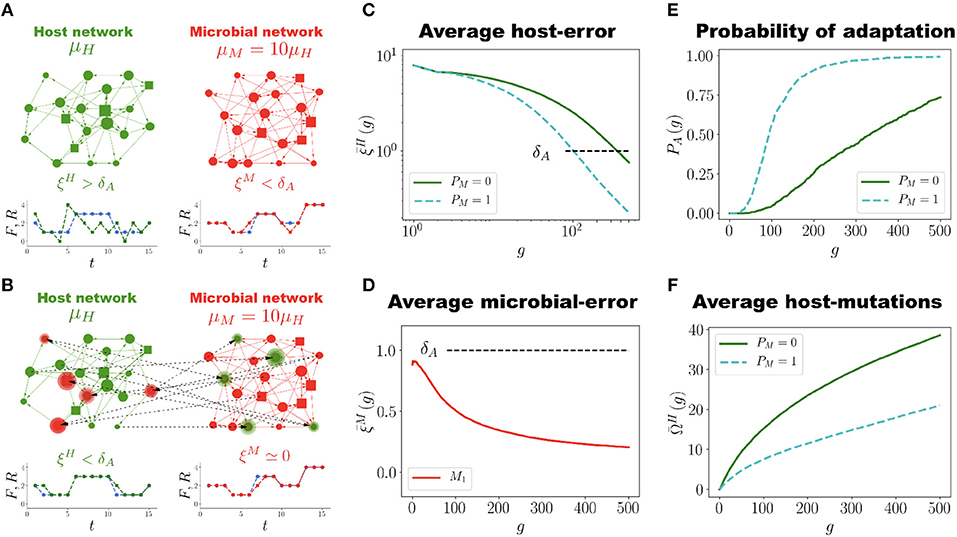
AF: We first represented microbes and hosts as independent dynamical gene networks, where genes interact through gene regulatory interactions. We then allowed mutation to occur at different rates. Evolution and adaptation were simulated through the requirement of specific collective “functional” tasks. These mutations can either modify the way a gene regulates another, or change the target of the regulatory interaction.
Since the premise is that the holobiont is a collection of interacting networks, a mutation can make a microbial gene become regulated by a host gene and vice versa. We took into account different approximate average timescales at which mutations occur in microbial and host networks. The performance of both hosts and microbes was measured independently by how well its gene expression can match a predetermined function assigned to each network.
What was your mathematical model able to predict?
AF: Apart from predicting dysbiosis when co-evolved networks are disconnected (simulating, for example, the action of antibiotics), one of the interesting predictions of our model is that when there are too many microbial communities contributing to the same host´s task, the task is often poorly performed – think of too many people trying to change a car’s tire.
This happens in spite of the fact that, in our model, a single microbial network contributing to a host´s task can significantly improve its performance.
However, when specific niche constraints (either territorial or metabolic) are introduced, allowing only a few microbial networks to contribute to a given task, the overall performance of the host is rapidly improved in comparison with a host without the support of microbial networks.
Host and Microbial Network Coevolution:
How do you think your model might be able to advance research on the human microbiome and health?
AF: One interesting aspect is to find and characterize the microbiome niches (which may not necessarily be anatomically separated), where only a subset of microbe species are normally involved. If too few or too many microbe species participate, this could lead to a dysbiotic state.
The specificity of microbial niches is crucial for future treatment of pathologies. In general terms, it now seems clear that considering the health of our microbial communities as an essential part of our well-being may lead to important discoveries in health and other human traits.
We believe that mathematical research, including time-series, network,and complexity science approaches will help shed light into the many different ways our microbiome interacts and has evolved with our bodies, as there is probably no linear relation between microbial diversity, abundances, and health.
How will you continue to improve on your model?
AF: At this stage, our paper may be considered as a basic framework for the description and simulation of the adaptation and heritability of our internal ecosystem. We are currently investigating, among other things, the effect of changing environments, modelled by evolving tasks for our networks.
The question of heritability of our microbiome is also a major issue to be addressed. For example, we plan to investigate the role of the so-called “core microbiome” (the microbes or microbial functions essential for our well being). We believe the model can be incrementally improved and made to better reflect specific situations, such as our microbiome’s role in antibiotic resistance, obesity, or other pathologies.
What do you think is ahead for mathematical modelling of the holobiont?
AF: I believe several different approaches will be necessary. On the one hand, new experimental methods and discoveries constantly occur, which will surely give rise to new theoretical ideas, which will have predictions, and so on, hopefully creating a virtuous circle – as in all good science.
With that said: our Boolean network approach has proved very useful in other areas, so we strongly believe it may provide a significant path for inquiry and modeling.


