Abstract
Knowledge about the human microbiome is requiring scientists to re-define the relationship between microbes and disease, going beyond Koch’s Postulates and incorporating concepts such as holobiont – understanding human physiology as the sum of the human body plus its microbiome– and pathobionts—microbes that cause disease only when certain conditions are met. Microbiome-based diagnostics are tools that allow detection of, and/or making prognoses on, human disease using microbial signatures of different types. Examples exist of potentially useful microbiome measurements at a disease site, or at a body site distant from the disease site. Causality between microbial signatures and disease is difficult to glean, yet in some cases microbiome signatures are useful diagnostically despite having no known causal relationship with the disease or condition in question. Mechanistic connections to disease are an active area of scientific inquiry. Companies globally are currently pursuing microbiome-based diagnostics or companion diagnostics for clinical use in a variety of diseases and conditions.
Introduction
For the past few centuries, by far the best-known microorganisms have been pathogens. In the early days of modern microbiology, the German physician Robert Koch established his Postulates, the criteria that have governed the basics of infectious diseases and the determination of a microorganism as an infectious agent for over a century. These rules are as follows:
- The microorganism must be found in all organisms suffering from the disease, but should not be found in healthy organisms.
- The microorganism must be isolated from a diseased organism and grown in pure culture.
- The cultured microorganism should cause disease when introduced into a healthy organism.
- The microorganism must be reisolated from the inoculated, diseased experimental host and identified as being identical to the original specific causative agent.
Today, having more detailed knowledge of host bacterial communities, we know that these postulates do not constitute the only possible relationship between microorganisms and disease in a host. Although these principles are still relevant, several updates have been proposed throughout the years,for instance, around the capacity of the microorganism to be grown in pure culture, and around the concept of opportunistic infections. Increased knowledge about a range of human afflictions—both infectious diseases and chronic conditions—has revealed that conceptualizations of the relationship between microbes and disease may need to be revisited.
The name microbiome-based diagnostics is given to the tools that allow detection of, and/or making prognoses on, human disease using microbial signatures. The main difference between these tools and the classic microbiological tests for infectious diseases is that microbiome diagnostics usually do not only aim at confirming the presence of a given microorganism as an infectious agent, but rather identify microbial signatures (e.g. presence, absence, or abundance of certain microbes and/or microbial activities) of different types (e.g. DNA, RNA, proteins or other metabolites), alone or in combination with other biomarkers (e.g. human genetics or metabolites), to detect disease and/or make prognoses. They are the result of the qualitative and quantitative integration of trans-kingdom, multi-omics biomarkers.
Microbiome diagnostics are based on emerging concepts in modern microbiology, some of which challenge Koch’s Postulates.
First, these new diagnostics must incorporate the concept of pathobionts, defined as symbiotic microorganisms that are able to promote pathology when specific conditions occur in the host: e.g. a certain genetic, environmental, or microbial context. Recently, some scientists have advocated for the substitution of this notion with the one of Pathogenic Potential (PP) of the human microbiota [1][2][3], and other researchers propose the entire dysbiotic microbiota (i.e. a network of pathobionts) should be regarded as an infectious agent [4]. In any case, what these ideas capture is that two people harboring similar microbial species or profiles in their respective microbiomes may have completely different reactions to it. The idea of pathobionts has led to the suggestion of revisiting the definition of noncommunicable diseases [5], as some of them may be transmitted through microbes, since pathobionts or components of dysbiotic microbiotas may be transferred from one individual to another through various means. In support of this notion, animal studies have shown that multiple traits or conditions can be transferred through fecal transplant, ranging from obesity [6][7][8][9], to altered behavior [10], even between different species [11].
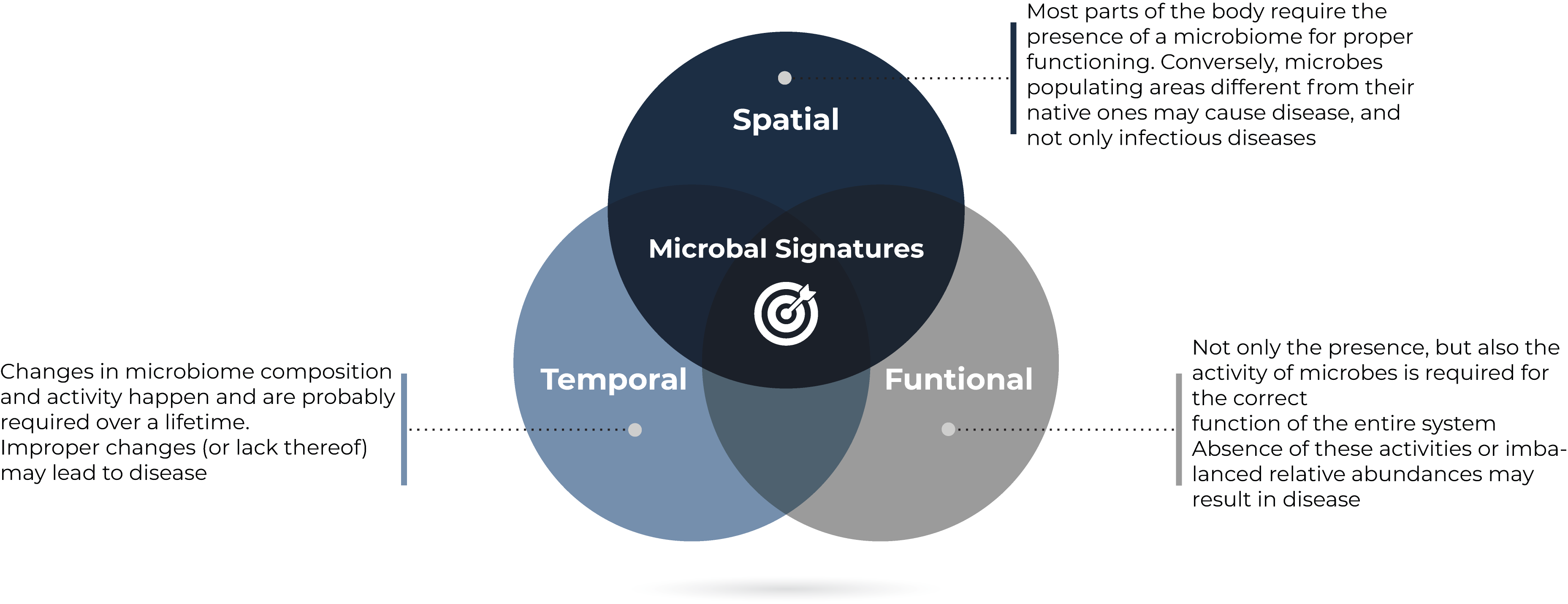
The second main concept to take into account for understanding microbiome diagnostics is the holobiont. Coined by Lynn Margulis in 1991 [12], it recasts the individual as a superorganism, or a community comprising the host and its symbiotic microbes, including the intracellular ones. This concept was reinforced in 2008, when Eugene Rosenberg and Ilana Zilber-Rosenberg proposed the hologenome theory of evolution [13], which considers the sum of the genetic information of the host plus its microbiome as the unit of natural selection. What these theories defend is that a proper fit between hosts and their microbiomes, optimized throughout eons of evolution, is required for the proper functioning of the system. If this system is disrupted, disease may arise.
Understanding individuals (human or otherwise) as holobionts has further implications for microbiome diagnostics. First, that not only the presence, but also the activity, of symbiotic microbes is required for the correct functioning of the entire system. And second, that microbes may have determined locations within the body in which their functions are required, and outside which their presence or activity may be detrimental (translocation). There are well-known opportunistic infections caused by gut or skin commensals; and an example of the deleterious effects of the translocation of the oral bacterium Porphyromonas gingivalis is discussed below.
Conversely, this idea implies that the absence of certain microbes, their metabolic activities or interactions with the host in sites where they should exist, may have negative consequences. A clear (albeit extreme) example is the multiple developmental problems observed in germ-free animals.
Lastly, the holobiont implies that this relationship between host and microbes may not only have a spatial component, but also a temporal one. Good examples of this are the evolution of the gut microbiome from birth to adulthood of a single host; and also the knowledge that an inverse relationship exists between H. pylori early-life colonization of the stomach and allergies, while a direct relationship exists between these same bacteria in adults and gastric cancer.Furthermore, modern techniques are enabling for the identification of more precise implications of the changes of the microbiome over time. For instance, abnormal daily oscillations in the composition of the gut microbiome may be a tool to predict the risk of type 2 diabetes [14].
1. Microbiome signatures in disease
In multiple human conditions initially believed to be independent from microorganisms or their metabolism [15], countless studies [16] have now linked dysfunction with microbiome abundance and/or activity profiles at the disease site. For example, specific microbial signatures have been found in the esophagus of individuals with esophageal cancer [17] in the stomach of gastric cancer patients [18], in the mouth affected by oral squamous cell carcinoma [19][20] in the lungs of people with COPD [21] or lung cancer [22] and in the gut in IBD-affected persons [23][24].
On some occasions, these microbial signatures not only vary with the presence of the disease but may also predict disease onset, remission, progression or the efficacy of certain treatments. For instance, the lung and sputum microbiomes of people with COPD change in connection with exacerbations [25] and intestinal and stool microbes may change before flares of IBD [26].
Intriguingly, also off-disease-site microbiomes seem to change in certain health ailments, like the changes observed in the throat microbial community of patients with schizophrenia [27] or the intestinal microbiome in Amyotrophic Lateral Sclerosis [28]. Sometimes these off-site microbiomes may be a good proxy to disease site changes and may be helpful for prognoses of disease course, such as the gut microbiome in pancreatic cancer [29] or NAFLD and cirrhosis [30]. However, distant microbial communities have proven helpful in predicting treatment success, as in the case of intestinal microbiome in melanoma therapy[31][32] as well as in many other immunotherapies for cancer. In medical conditions where the disease is not located on a specific site, changes in the gut microbiome also appear repeatedly—such as in obesity and diabetes [33] —indicating that it is an important site of microbiome activity. Although most of the current evidence has been generated onbacteria,research shows that other members of the microbiome, such as viruses, may also harbor signatures useful in diagnosing or prognosing disease[34].
Not only microorganisms themselves, but also their metabolites,may also show disease-specific signatures. The blood is becoming known as a source of information about the microbes in the body, particularly those in the gut. It should not be forgotten that an estimated 15-30% of the metabolites in human blood are derived from microbes, especially from those dwelling in the intestine [35] and that the microbiome is indeed one of the main determinants of the composition of the blood metabolome [36]. In recent years, a number of studies analyzing blood for microbial signatures have been published. For example, Poore et al. recently [37] found they could discriminate between samples from healthy individuals and those from patients with melanoma, and prostate and lung cancer with fair accuracy using plasma-derived, cell-free microbial nucleic acids [38]. Also looking at cell-free microbial DNA, Dzutsev & Trinchieri [39] could distinguish patients with cancer from individuals without cancer and, moreover, identify the tumor type. Further study may expand this in the years ahead.
Lastly, it may also be surprising that non-infectious microbial signatures may also help, not in diagnosing infections per se, but indeed in predicting certain aspects of classicinfectious diseases. For instance, specific gut microbial characteristics have been linked to predisposition to severe COVID-19 [40]. Also, and despite its possible status as a pathobiont disease, researchers have shown that Clostridioides difficile infection (CDI) recurrence and treatment response can be accurately predicted by looking at the broader microbial community in the intestine and not exclusively attending at the infectious agent itself [41].
2. A chicken-or-egg situation
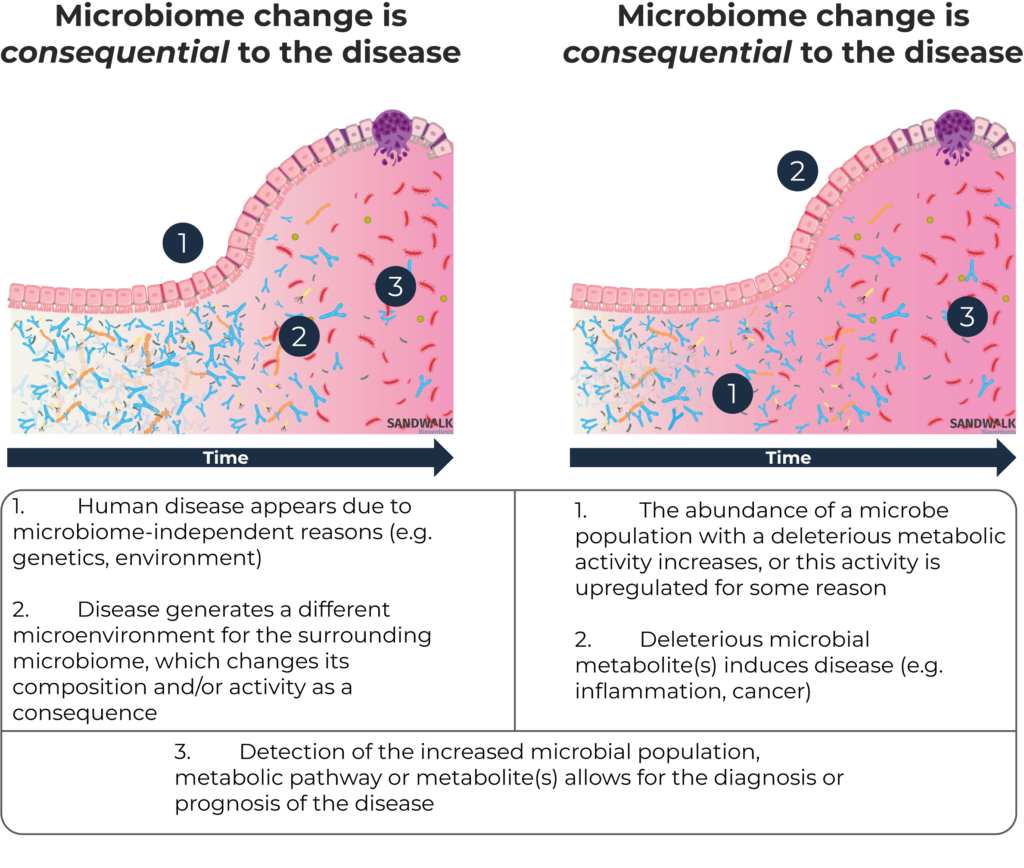
The cause vs.consequence question constantly arises in studies around microbiome and disease. The degree to which observed microbial changes are either the cause of the disease, or a consequence of the disease processes that occur regardless, is largely unknown; authors have recommended caution in assuming causality since the early days of microbiome research [42].
On some occasions, where the microbiome-independent etiology of the disease is well known, detected microbial signatures are clearly a consequence of the disease. Cystic fibrosis (CF) is a disorder which affects many organs in the body—most notably the lungs, but also the pancreas, liver, kidneys and intestine [43] as people with this condition produce bodily secretions thicker than normal. It is a genetic disease and its molecular bases are exceptionally well described – over 1,500 different mutations in the cystic fibrosis transmembrane conductance regulator (CFTR) gene, with one particular mutation responsible for the vast majority of cases [44]. The affected individuals have a life expectancy shorter than normal and experience multiple symptoms throughout life, especially gastrointestinal manifestations and lung infections. For those with CF, multiple studies on microbiome composition of different body sites, mainly intestine and lungs, have been published [45]. These studies consistently show microbiome aberrations compared to disease-free individuals; however, these modifications can be originally attributed to the altered environment generated by the thicker body secretions. Therefore, these microbiome signatures are clearly not the cause of the disease, but rather a consequence. Since established methods for diagnosis of this condition already exist, the microbiome data is not useful for the diagnosis of CF. The data may, though, be useful for its prognosis as the altered microbiome may be directly or indirectly behind some of the symptoms of the disease, or even behind CF’s most serious consequences.
Microbiome alterations that are consequential to diseases that have a microbiome-independent onset are sometimes regarded as merely anecdotal and useless in diagnosis or drug development. However, sometimesthey may be very useful as a means for non-invasive disease detection or inexpensive mass screening as an alternative to, or even as a more reliable option compared to existing diagnostics. For example, researchers in China recently suggested that the alteration in the composition of the gut microbiome after stroke may be a good predictor of post-stroke cognitive impairment (PSCI) [46]. The precise pathophysiology of PSCI is not fully understood, so no tools are currently available to predict its onset. According to this research, gut microbiome signatures may become one of the few biomarkers to look at in this condition. Other diseases such as Alzheimer’s where no powerful diagnostics exist may also benefit from microbiome interrogation, according to recent research [47].
Despite the potential usefulness of indirect and/or consequential microbiome signatures in disease diagnosis or prognosis, especially where other biomarkers may not exist and for non-invasive, fast, and inexpensive screening, the real goal for researchers is to find microbiome signatures that may be causative of disease. Finding microbiome profiles that may lead to the onset of a disease would not only aid in its diagnosis or early detection, but also potentially lead to the discovery of new druggable targets for its prevention or treatment.
3. The search for mechanistic connections
To the extent that disease-causing microbiome alterations exist in humans, discovery of these causal microbiome signatures is the holy grail of microbiome diagnostics and drug development. Nonetheless, the mechanisms behind these relationships are usually tremendously complex and involve not only the microbes themselves, but also their metabolites and aspects of humangenetics and the immune, endocrine, and nervous systems.
The capacity of some infectious microorganisms to cause cancer is well known, as infections by H. pylori, Human Papilloma Viruses, Hepatitis B and C Viruses, Epstein-Barr Virus, the Human Immunodeficiency Virus and Human Herpes Viruses are collectively responsible for around 18% of all cancer cases worldwide [48]. However, the role of non-infectious microbes in cancer onset and progression is only starting to being explored.
Fusobacterium nucleatum is a naturally-occurring member of the microbiome of the human mouth and intestine that may act as an opportunistic pathogen, causing infections. Over the last decade, multiple studies have shown that it is more abundant in the microbiome of CRC patients [49] and that its enrichment is correlated with poor prognosis in this disease [50][51]. This placed F. nucleatum as a candidate for a cancer-causing bacterium. So far, a number of mechanisms have been suggested for carcinogenesis and poor prognosis mediated through F. nucleatum, although these relationships are not yet completely understood [52]. It is therefore unclear why some individuals develop F. nucleatum-mediated CRC, what other factors are involved and whether this bacterium alone, and if so in which abundance, is enough to generate CRC. To add more unknowns, Fusobacterium has been detected not only in primary CRC but also in distant metastases, and the bacterium actually persisted when tumors were transplanted several times in immunocompromised mice. In one study, antibiotic treatments that eliminated fusobacteria decreased the ability of tumors to grow [53][54]. More detailed insights have been generated around the mechanisms by which other bacteria may lead to carcinogenesis.
Escherichia coli is a common and very well studied commensal species of the human intestine. Some of its strains have genetic elements in their chromosome, called pks islands, that code for the machinery required to produce a molecule called colibactin. Colibactin has been shown to accelerate tumor formation in animal models [55] by means of causing two very specific types of DNA mutations. These exact mutations are present in around 8% of colorectal cancer metastases, 3% of urinary tract cancers, and 2% of head and neck tumors where E. coli may be involved. Furthermore, pks+E. coli is found in 67% of people with colorectal cancer [56], which may suggest this mechanism is implicated in the development of the malady. Also, the presence of pks+E. coli in the fecal microbiome of IBD patients is threefold higher than in healthy individuals [57], which is being hypothesized as one of the reasons, alongside immune factors, for IBD patients having significantly higher chances of developing CRC than the general population [58]. It is known that E. coli is overrepresented in the fecal microbiome of IBD patients and it has been suggested that this microbe may partially be causative of the development of IBD [59]. Analogous CRC-promoting effects have recently been attributed to a different gut microbiome derived molecule, gallic acid [60].
As discussed above, a significant share of the metabolites in our blood are derived from microbes, so systemically-acting gut microbial metabolites may also be useful biomarkers for diagnosis. For example, Menni et al. reported that microbiome metabolites found in blood are significantly altered in patients with type 2 diabetes [61]. Research also shows that up to 95% of the metabolites in feces are synthesized by gut microbes [62], and as one example, a recent report by another group showed that microbial metabolites in feces are significantly related to the risk of dementia [63].
In addition to modification of the local environment by bacteria, and distant effects through metabolites, microbial translocation is another mechanism by which microorganisms may lead to non-infectious disease.
The relationship between periodontitis and cardiovascular disease has been known for long [64][65]. Their association was thought to be mediated through a systemic inflammation, which manifested through gum swelling but also through atherosclerosis. Some studies looked at translocation of gum microbes, such as Porphyromonas gingivalis, to the bloodstream and generation of antibodies as a potential link [66]. Later, an association between IgG against P. gingivalis and cognitive impairment, independent from cardiovascular factors, was established [67]. In 2016, researchers discovered that amyloid, the protein whose accumulation in the brain is believed to lead to Alzheimer’s Disease (AD), may act as a defense against bacterial infections in the brain [68]. Later a rodent study found that P. gingivalis can actively travel from the mouth to the brain and invade it [69]and that some enzymes that this microbe uses to feed on human tissue have been found in up to 99% of brains of patients with AD [70], whereas the brains from people without AD have lower levels of P. gingivalis accumulation. These observations have led to an emerging theory that P. gingivalis translocation to the brain may trigger the release of amyloid to contain the infection, which in turn causes an excessive immune response that ends up killing neurons instead of protecting them, causing AD [71].
Bacteria were first detected within human tumors more than 100 years ago, but the characterization of the in-tumor microbiome has remained challenging due to its small biomass. Improvement of techniques has allowed for these kinds of analyses, and a recent publication reported that bacteria can be found in tumors from all cancer types. For instance, more than 60% of bone, breast and pancreatic tumors and 14% of melanomas tested positive for bacterial DNA belonging to 528 different species. Bacteria in tumors are mostly intracellular and are present in both cancer and immune cells, and although previous studies found a very low amount of bacteria in tumors, modern analyses were able to detect up to 40 bacteria per human cancer cell [72]. Even though this is very unclear, bacteria are thought to colonize malignant tissues due to the anaerobic niche tumors generate, although other factors may play a role in their preference for certain types of tumors. For example, lung tumors in smokers tended to contain bacteria that break down tobacco chemicals [73].
Independently from their role in the appearance of cancer, intra-tumoral bacteria seem to have a profound effect on cancer progression and therapeutic outcomes. In fact, Hermida et al. found that tumor microbiome is better than tumor transcriptome in predicting chemotherapy response [74].
Presence of some bacteria in tumors has been linked to both positive and negative therapeutic outcomes. For example, it is known that tumor infection of some microbes is capable of killing cancer cells through various mechanisms [75] and that the presence of a broader diversity of microorganisms in-tumor is linked to better disease prognosis in pancreatic cancer, probably through an activation or attraction of immune cells to the tumor [76]. On the other hand, the opposite has been described in lung cancer [77]. It is also known that certain bacteria are capable of enzymatically inactivating anticancer drugs on-site, like tumor-associated proteobacteria do with the cytotoxic drug gemcitabine [78] limiting the efficacy of the intervention. Yet locally-acting microbial metabolites have also been linked to an increased efficacy of cancer treatments. Working with a mouse model of CRC, Mager et al. showed that the gut microbiome may positively affect the efficacy of immune checkpoint inhibitors through the production of the nucleoside inosine [79].
Therefore, when these relationships and mechanisms are better understood, there may be room for microbial analyses of solid or liquid biopsies alongside genetic and pathological anatomy tests, in order to fully characterize the malignancy and its microenvironment, and to help design tailored therapeutic interventions.
4. Companies pursuing microbiome biomarkers
All the links between microbiome composition and/or activities and human disease have captured the attention of not only academics and funders but also of industry. A number of companies have been founded around technologies that aim at uncovering and validating the mechanisms that underlie observed relationships, and some of the major deals with big pharma involve the use of these platforms to find new drugs.
In June 2020, Takeda and Debiopharm (Lausanne, Switzerland) disclosed their licensing deal to develop novel microbiome therapeutics for IBD and other gastrointestinal disorders by employing Debiopharm’s discovery platform for narrow-spectrum microbiome modulation agents [80]. This deal also shows Takeda’s interest in microbiome technologies (between 2017 and 2020 it has signed deals with NuBiyota, Finch Therapeutics and Enterome) and also their position as a leading company in gastroenterology after their €53Bn acquisition of Shire Pharmaceuticals in 2019 [81].
More recently, BiomX, Inc. (Ness Ziona, Israel; NYSE: PHGE) entered a collaboration with Boehringer Ingelheim to utilize BiomX’s microbiome-based biomarker discovery platform to identify biomarkers associated with patient phenotypes in IBD [82].
The microbiome is indeed a potential source of an immense amount of new druggable targets. However,we will focus on groups developing diagnostics and companion diagnostics. These may in turn, some day, become drug targets theselves.
Microbiome-based diagnostics
Colorectal cancer (CRC) is one of the top causes of cancer mortality in the world. Diagnosis of CRC is performed or confirmed via colonoscopy, which is an invasive and expensive technique that is not free of risk. A common, non-invasive, safe and inexpensive screening test for CRC is based on the human hemoglobin immunochemical based fecal occult blood test, which consists in the detection of blood in stool. This is a very useful technology for population screening purposes, but it exhibits a false-positive rate of around 40% [83], such false positives need to be confirmed by colonoscopy. This is one reason why stool microbial signatures are being sought as an alternative to, or additional screening method for, CRC [84]. As they may capture many more factors than immunochemical techniques, these microbiome-based tests could have a much better specificity profile, even if the detected signatures are the microbial changes resulting from the root cause of intestinal bleeding in CRC, and are therefore clearly consequential and indirect rather than causative of the disease. In fact, some companies believe these techniques may become so sensitive that they may substitute for colonoscopy in this context, as fecal microbiome changes may also reveal pre-cancerous, pre-bleeding stages. Furthermore, these tools may be based on PCR technology rather than on sequencing, making them faster and cheaper. Some companies active in this segment are Pescient Metabiomics, LLC (Carlsbad, CA, USA), GLC Biotechnology, Inc. (Hudson, OH, USA), ZMD Group (Netivot, Israel), GoodGut S.L. (Barcelona, Spain) and Bio-Me A/S (Oslo, Norway).
Outside CRC, Teraomics S.L. (Alicante, Spain) is validating its diagnostic tool for detection and progression of head and neckcancer based on samples of saliva.
Beyond oncology, a number of companies are developing diagnostics for diseases of the intestine through stool microbiome analysis. That is the case for IBD, for which GoodGut and AlphaBiomics Ltd (London, UK) are creating applications, and the non-celiac gluten sensitivity test by Amyra Biotech AG (Basel, Switzerland). Stool is also being employed by Teraomics to validate its early discoveries that intestinal microbiome signatures may be linked to the risk of progression and may help in the prediction of treatment response in multiple sclerosis.
Apart from diseases of the gastrointestinal tractand stool samples, ARTPred B.V. (Hoofddorp, Netherlands) are focusing on the role of the female reproductive tract microbiome on the outcomes of in vitro fertilization treatments, given the connections uncovered between the composition of this microbiome and fertility. Microbiome-focused liquid biopsy and analysis of cancer tissue associated microbes to detect and discriminate between types of cancer is an emerging technology being developed by Micronoma (San Diego, CA, USA), while The BioCollective (Denver, CO) and Ardigen (Kralow, Poland) have established a partnership to identify gut microbiome biomarkers to detect early-onset Parkinson’s Disease and its progression.
Endobios (Sintra, Portugal) are going a step further and developing diagnostics and associated therapies by studying endosymbiotic relationships.
Companion diagnostics
One of the applications of microbiome biomarker discovery that is becoming most relevant over the past few years is companion diagnostics. Some companies are applying their microbiome-mining platforms to identify signatures in the microbiota to predict efficacy of established drugs, many of them in close collaboration with major pharmaceutical companies, whereas others are trialing their microbiome therapeutics as adjuvant therapies with medications, mainly for autoimmune diseases [85].
For instance, Second Genome (Brisbane, CA, USA) and Gilead (Nasdaq: GILD) signed an agreement in 2020 by which Second Genome’s Microbiome Analytics Platform™ will be employed to identify biomarkers associated with clinical response to Gilead’s investigational medicines in IBD. Under the terms of the agreement, Second Genome will receive from Gilead €32M in an upfront payment, and up to €256 million in success-based milestones [86]. This deal is particularly paradigmatic as it reinforces the trend of increasing collaboration between big pharma and microbiome startups to identify adjuvant therapies and companion diagnostics for non-microbiome-based drugs, discussed above. It also highlights Gilead’s relatively recent expansion into immune diseases and oncology, which has also materialized this year with other strategic collaborations outside microbiome with Tango Therapeutics (August 2020), Jounce Therapeutics (September 2020) and their acquisition of Forty-Seven, Inc. (€4Bn, March 2020) and Immunomedics (€18Bn, September 2020).
In the field of oncology, Persephone Biosciences, Inc. (San Diego, CA, USA) are engaged in an observational trial (ATALANTA, NCT04291755) to identify microbial biomarkers in blood, urine, and stool predictive of the efficacy of Keytruda®(pembrolizumab) in lung and colorectal cancers. Keytruda®’s maker, Merck & Co. (NYSE: MRK) is one of big pharma’s most active players in researching the interaction between monoclonal antibodies and microbiome [87]. Persephone recently announced the initiation of an additional observational study (ARGONAUT, NCT04638751) for the development of companion diagnostics in lung, breast, colorectal and pancreatic cancers being treated with immunotherapy [88].
Outside these applications, New Biotic, Inc. (Irvine, CA, USA), which have developed a microbiome-based therapy for Amyotrophic Lateral Sclerosis, are simultaneously working on a companion diagnostic to predict therapeutic efficacy and disease progression, based on the detection of serum glutamate levels and glutamine synthetase activity in the intestine. Also, Siolta Therapeutics (San Carlo, CA) are developing a tool in collaboration with Bio-Me (Oslo, Norway) for patient stratification in upcoming clinical trials with a live biotherapeutic product for asthma [89].
The image below shows a breakdown of all the diagnostics and companion diagnostics development programs in the industry, segmented by their application [90]. Relationships between the microbiome and the immune system, especially around cancer and auto-immune diseases, capture most of this market. Non-classical ways to detect, stratify or prognose infectious diseases rank third.
5. Future challenges
The main challenge facing microbiome diagnostics is validation, as microbial signatures change in response to many types of factors, making it difficult to establish clear-cut connections between disease course / onset and the microbiome. Furthermore, we are probably still far from finding out what the microbiome should look like (eubiosis), and conversely what the opposite (dysbiosis) means. This is always an important limitation when trying to define health and disease. However, steady efforts and advances are being made [91].
Also, in order to validate diagnostics biomarkers or druggable targets based on the microbiome, extremely large datasets and patient populations are required. After all, the Human Microbiome Project ran for 10 years and was funded with €145M; and despite its notable achievements, it was merely a first step and did not definitively characterize what it means to have a ‘healthy’ human microbiome. Lower costs for sequencing and further sophistication in big data analysis, artificial intelligence, and machine learning tools will be crucial for the continued development of this sector.
All in all, the microbiome is an extremely exciting and promising area for the generation of new diagnostics, prognostics and therapeutics. The early scientific studies provide an indication that most of the opportunities in this field are still untapped, with the complex interplay between elements of the human holobiont, resulting in states of health and disease, only starting to be uncovered.
- Ochum, L and Stecher, B ‘’Label or Concept – What Is a Pathobiont?’’. Trends in Microbiology 28:10 (October 2020): 789-792
- Chow, J and Mazmanian, S. ‘’ A Pathobiont of the Microbiota Balances Host Colonization and Intestinal Inflammation’’. Cell Host & Microbe 7 :4, (22 April 2010) : 265-276
- Mazmanian, S., Round, J. & Kasper, D. ‘’A microbial symbiosis factor prevents intestinal inflammatory disease’’. Nature 453, 620–625 (2008). https://doi.org/10.1038/nature07008
- Finlay, B. ‘’Are noncommunicable diseases communicable?’’. Science 17 Jan 2020: Vol. 367, Issue 6475, pp. 250-251. DOI: 10.1126/science.aaz3834
- Gosalbez, L. La microbiota humana como estrategia farmacológica en el entorno regulatorio europeo. http://repositorio.ucam.edu/handle/10952/2496.
- Bäckhed, F et al. ‘’ The gut microbiota as an environmental factor that regulates fat storage’’. PNAS November 2, 2004 101 (44) 15718-15723; https://doi.org/10.1073/pnas.0407076101
- Ellekilde, M., Selfjord, E., Larsen, C. et al. Transfer of gut microbiota from lean and obese mice to antibiotic-treated mice. Sci Rep 4, 5922 (2014). https://doi.org/10.1038/srep05922
- Ridaura, V. et al. “Gut microbiota from twins discordant for obesity modulate metabolism in mice.” Science (New York, N.Y.) vol. 341,6150 (2013): 1241214. doi:10.1126/science.1241214
- Leocádio, P. et al ‘’ Obesity: More Than an Inflammatory, an Infectious Disease?’’. Front. Immunol., 14 January 2020 | https://doi.org/10.3389/fimmu.2019.03092
- D’Amato, A. et al ‘’Faecal microbiota transplant from aged donor mice affects spatial learning and memory via modulating hippocampal synaptic plasticity- and neurotransmissionrelated proteins in young recipients’’. Microbiome (2020) 8:140 https://doi.org/10.1186/s40168-020-00914-w
- De Palma, G. et al ‘’Transplantation of fecal microbiota from patients with irritable bowel syndrome alters gut function and behavior in recipient mice’’. Sci Transl Med : 2017 Mar 1;9 (379):eaaf6397. doi: 10.1126/scitranslmed.aaf6397. PMID: 28251905.
- Lynn Margulis, L. & Fester, R. Symbiosis as a Source of Evolutionary Innovation: Speciation and Morphogenesis, Amherst, MIT Press, 1991
- Zilber-Rosenberg, I and Rosenberg, E. ‘’Role of microorganisms in the evolution of animals and plants: the hologenome theory of evolution’’, FEMS Microbiology Reviews, Volume 32, Issue 5, August 2008, Pages 723–735, https://doi.org/10.1111/j.1574-6976.2008.00123
- Reitmeier, S et al, ‘’Arrhythmic Gut Microbiome Signatures Predict Risk of Type 2 Diabetes’’ Cell Host & Microbe, 28: 2, 12 August 2020, pp. 258-272, DOI:https://doi.org/10.1016/j.chom.2020.06.004
- Durack, J. and S. Lynch. “The gut microbiome: Relationships with disease and opportunities for therapy.” The Journal of experimental medicine vol. 216,1 (2019): 20-40. doi:10.1084/jem.20180448
- Ibid
- Lopetuso, L et al. “Esophageal microbiome signature in patients with Barrett’s esophagus and esophageal adenocarcinoma.” PloS one vol. 15,5 e0231789. 5 May. 2020, doi:10.1371/journal.pone.0231789
- Wang, Z. et all ‘’ Changes of the Gastric Mucosal Microbiome Associated With Histological Stages of Gastric Carcinogenesis’’. Front. Microbiol., 29 May 2020, https://doi.org/10.3389/fmicb.2020.00997
- York, A. ‘’P. gingivalis drives carcinoma progression’’. Nat Rev Microbiol 18, 674 (2020). https://doi.org/10.1038/s41579-020-00466-8
- Minarovits, J. ‘’ Anaerobic bacterial communities associated with oral carcinoma: Intratumoral, surface-biofilm and salivary microbiota’’ Anaerobe, 2020, 102300, https://doi.org/10.1016/j.anaerobe.2020.102300.
- Wang, L. et al. “Role of the Lung Microbiome in the Pathogenesis of Chronic Obstructive Pulmonary Disease.” Chinese medical journal vol. 130,17 (2017): 2107-2111. doi:10.4103/0366-6999.211452
- Ramírez-Labrada, A. et al ‘’ The Influence of Lung Microbiota on Lung Carcinogenesis, Immunity, and Immunotherapy’’ Trends in Cancer, 6 :2, 1 February 2020, p. P86-97, https://doi.org/10.1016/j.trecan.2019.12.007
- Alam, M.T., Amos, G.C.A., Murphy, A.R.J. et al. ‘’Microbial imbalance in inflammatory bowel disease patients at different taxonomic levels’’. Gut Pathog 12, 1 (2020). https://doi.org/10.1186/s13099-019-0341-6
- Galipeau, H. et al ‘’Novel fecal biomarkers that precede clinical diagnosis of ulcerative colitis’’ Gastroenterology, 10 December 2020, https://doi.org/10.1053/j.gastro.2020.12.004
- Wang, Z. ‘’Lung microbiome dynamics in COPD exacerbations
- Lloyd-Price, J., Arze, C., Ananthakrishnan, A.N. et al. ‘’Multi-omics of the gut microbial ecosystem in inflammatory bowel diseases’’. Nature 569, 655–662 (2019). https://doi.org/10.1038/s41586-019-1237-9
- Castro-Nallar, E. et al. “Composition, taxonomy and functional diversity of the oropharynx microbiome in individuals with schizophrenia and controls.” PeerJ vol. 3 e1140. 25 Aug. 2015, doi:10.7717/peerj.1140
- Zeng, Q., Shen, J., Chen, K. et al. ‘’The alteration of gut microbiome and metabolism in amyotrophic lateral sclerosis patients’’. Sci Rep 10, 12998 (2020). https://doi.org/10.1038/s41598-020-69845-8
- Riquelme, E. et al. “Tumor Microbiome Diversity and Composition Influence Pancreatic Cancer Outcomes.” Cell vol. 178,4 (2019): 795-806.e12. doi:10.1016/j.cell.2019.07.008
- Gyu Oh, T. et al ‘’ A Universal Gut-Microbiome-Derived Signature Predicts Cirrhosis’’, Cell Metabolism, 32 :5, 3 November 2020, p. 878-888, DOI:https://doi.org/10.1016/j.cmet.2020.06.005
- Gopalakrishnan, V. et al ‘’Gut microbiome modulates response to anti–PD-1 immunotherapy in melanoma patients’’, Science, 05 January 2018, 359:6371, pp. 97-103, DOI: 10.1126/science.aan4236
- Chaput, N. ‘’ Baseline gut microbiota predicts clinical response and colitis in metastatic melanoma patients treated with ipilimumab’’, Annals of Oncology, 28 : 6, June 2017, p. 1368-1379, https://doi.org/10.1093/annonc/mdx108
- Komaroff AL. ‘’The Microbiome and Risk for Obesity and Diabetes’’. JAMA. 2017;317(4):355–356. doi:10.1001/jama.2016.20099
- Lang, S. ‘’ Intestinal Virome Signature Associated With Severity of Nonalcoholic Fatty Liver Disease’’, Gastroenterology, 159 : 5, November 2020, p. 1839-1852, https://doi.org/10.1053/j.gastro.2020.07.005
- Visconti, A., Le Roy, C.I., Rosa, F. et al. ‘’Interplay between the human gut microbiome and host metabolism’’. Nat Commun 10, 4505 (2019). https://doi.org/10.1038/s41467-019-12476-z
- Bar, N., Korem, T., Weissbrod, O. et al. ‘’A reference map of potential determinants for the human serum metabolome’’. Nature 588, 135–140 (2020). https://doi.org/10.1038/s41586-020-2896-2
- Poore, G.D., Kopylova, E., Zhu, Q. et al. ‘’Microbiome analyses of blood and tissues suggest cancer diagnostic approach’’. Nature 579, 567–574 (2020). https://doi.org/10.1038/s41586-020-2095-1
- Ibid
- Dzutsev, A., Trinchieri, G. ‘’Microbial DNA signature in plasma enables cancer diagnosis’’. Nat Rev Clin Oncol 17, 453–454 (2020). https://doi.org/10.1038/s41571-020-0391-1
- Gou, W. ‘’Gut microbiota may underlie the predisposition of healthy individuals to COVID-19’’. medRxiv, 25 April 2020, https://doi.org/10.1101/2020.04.22.20076091
- Khanna, S. et al. “Gut microbiome predictors of treatment response and recurrence in primary Clostridium difficile infection.” Alimentary pharmacology & therapeutics vol. 44,7 (2016): 715-727. doi:10.1111/apt.13750
- Hanage, W., ‘’Microbiology: Microbiome science needs a healthy dose of scepticism’’ Nature, 512:7514,
- 20 August 2014
- O’Sullivan, B. ‘’Cystic fibrosis’’ The Lancet, 373:9678, 30 May 2009, p. 1891-1904, https://doi.org/10.1016/S0140-6736(09)60327-5
- Bobadilla, J.L., Macek, M., Jr, Fine, J.P. and Farrell, P.M. (2002), ‘’Cystic fibrosis: A worldwide analysis of CFTR mutations—correlation with incidence data and application to screening’’. Hum. Mutat., 19: 575-606. https://doi.org/10.1002/humu.10041
- Huang YJ, LiPuma JJ. ‘’The Microbiome in Cystic Fibrosis’’. Clin Chest Med. 2016 Mar;37(1):59-67. DOI: 10.1016/j.ccm.2015.10.003. Epub 2015 Dec 23. PMID: 26857768; PMCID: PMC5154676.
- Ling, Yi et al. “Gut Microbiome Signatures Are Biomarkers for Cognitive Impairment in Patients With Ischemic Stroke.” Frontiers in aging neuroscience vol. 12 511562. 23 Oct. 2020, doi:10.3389/fnagi.2020.511562
- Marizzoni, M. et al. ‘’Short-Chain Fatty Acids and Lipopolysaccharide as Mediators Between Gut Dysbiosis and Amyloid Pathology in Alzheimer’s Disease’’. 1 Jan. 2020 : 683 – 697.
- Parkin, D.M., ‘’The global health burden of infection‐associated cancers in the year 2002’’. Int. J. Cancer, 118: 3030-3044. https://doi.org/10.1002/ijc.21731
- Fukugaiti, M. H. et al. “High occurrence of Fusobacterium nucleatum and Clostridium difficile in the intestinal microbiota of colorectal carcinoma patients.” Brazilian journal of microbiology : [publication of the Brazilian Society for Microbiology] vol. 46,4 (2015): 1135-40. doi:10.1590/S1517-838246420140665
- Mima, K. et al. “Fusobacterium nucleatum in colorectal carcinoma tissue and patient prognosis.” Gut vol. 65,12 (2016): 1973-1980. doi:10.1136/gutjnl-2015-310101
- Yu, T. et al. “Fusobacterium nucleatum Promotes Chemoresistance to Colorectal Cancer by Modulating Autophagy.” Cell vol. 170,3 (2017): 548-563.e16. doi:10.1016/j.cell.2017.07.008
- Sun, C. et al. “The role of Fusobacterium nucleatum in colorectal cancer: from carcinogenesis to clinical management.” Chronic diseases and translational medicine vol. 5,3 178-187. 1 Oct. 2019, doi:10.1016/j.cdtm.2019.09.001
- Bullman, S. et al. “Analysis of Fusobacterium persistence and antibiotic response in colorectal cancer.” Science (New York, N.Y.) vol. 358,6369 (2017): 1443-1448. doi:10.1126/science.aal5240
- Dzutsev, A., Trinchieri, G., ‘’Microbial DNA signature in plasma enables cancer diagnosis’’. Nat Rev Clin Oncol 17, 453–454 (2020). https://doi.org/10.1038/s41571-020-0391-1
- Arthur, JC et al. “Intestinal inflammation targets cancer-inducing activity of the microbiota.” Science (New York, N.Y.) vol. 338,6103 (2012): 120-3. doi:10.1126/science.1224820
- Pleguezuelos-Manzano C. et al‘’Mutational signature in colorectal cancer caused by genotoxic pks+ E. coli’’. Nature. 2020 Apr;580(7802):269-273. doi: 10.1038/s41586-020-2080-8. Epub 2020 Feb 27. PMID: 32106218.
- Dubinsky V. et al ‘’Carriage of Colibactin-producing Bacteria and Colorectal Cancer Risk’’. Trends in Microbiology, 28:11, 1 November 2020, p. 874-876, DOI:https://doi.org/10.1016/j.tim.2020.05.015
- Clarke WT, Feuerstein JD. ‘’Colorectal cancer surveillance in inflammatory bowel disease: Practice guidelines and recent developments’’. World J Gastroenterol 2019; 25(30): 4148-4157, DOI: https://dx.doi.org/10.3748/wjg.v25.i30.4148
- Rhodes, JM. “The role of Escherichia coli in inflammatory bowel disease.” Gut vol. 56,5 (2007): 610-2. doi:10.1136/gut.2006.111872
- Kadosh, E., Snir-Alkalay, I., Venkatachalam, A. et al. ‘’The gut microbiome switches mutant p53 from tumour-suppressive to oncogenic’’. Nature 586, 133–138 (2020). https://doi.org/10.1038/s41586-020-2541-0
- Menni C. et al, ‘’Serum metabolites reflecting gut microbiome alpha diversity predict type 2 diabetes’’, Gut Microbes, Volume 11, 2020 – Issue 6, p. 1632-1642, https://doi.org/10.1080/19490976.2020.1778261
- Visconti, A., Le Roy, C.I., Rosa, F. et al. ‘’Interplay between the human gut microbiome and host metabolism’’. Nat Commun 10, 4505 (2019). https://doi.org/10.1038/s41467-019-12476-z
- Saji, N., Murotani, K., Hisada, T. et al. ‘’Relationship between dementia and gut microbiome-associated metabolites: a cross-sectional study in Japan’’. Sci Rep 10, 8088 (2020). https://doi.org/10.1038/s41598-020-65196-6
- Scannapieco FA, Bush RB, Paju S. ‘’Associations between periodontal disease and risk for atherosclerosis, cardiovascular disease, and stroke. A systematic review’’. Ann Periodontol. 2003 Dec;8(1):38-53. doi: 10.1902/annals.2003.8.1.38. PMID: 14971247.
- Janket SJet al. ‘’Meta-analysis of periodontal disease and risk of coronary heart disease and stroke. Oral Surg Oral Med Oral Pathol Oral Radiol Endod’’. 2003 May;95(5):559-69. doi: 10.1067/moe.2003.107. PMID: 12738947.
- Pussinen, P. et al ‘’Systemic exposure to Porphyromonas gingivalis predicts incident stroke’’. Atherosclerosis. 193:1, 1 July 2007, p. 222-228, DOI:https://doi.org/10.1016/j.atherosclerosis.2006.06.027
- Noble, J M et al. “Periodontitis is associated with cognitive impairment among older adults: analysis of NHANES-III.” Journal of neurology, neurosurgery, and psychiatry vol. 80,11 (2009): 1206-11. doi:10.1136/jnnp.2009.174029
- Kumar, D. et al ‘’Amyloid-β peptide protects against microbial infection in mouse and worm models of Alzheimer’s disease’’. Science Translational Medicine. 8:340, 25 May 2016, p. 340-372, DOI: 10.1126/scitranslmed.aaf1059
- Poole, S. et al. ‘Active Invasion of Porphyromonas Gingivalis and Infection-Induced Complement Activation in ApoE -/- Mice Brains’. 1 Jan. 2015 : 67 – 80.
- Dominy, S. et al ‘’Porphyromonas gingivalis in Alzheimer’s disease brains: Evidence for disease causation and treatment with small-molecule inhibitors’’. Science Advances. 5:1, 23 Jan 2019, DOI:10.1126/sciadv.aau3333
- Mackenzie, D. ‘’We may finally know what causes Alzheimer’s – and how to stop it’’. Health. 23 January 2019 ,updated 30 January 2019. https://www.newscientist.com/article/2191814-we-may-finally-know-what-causes-alzheimers-and-how-to-stop-it/
- Dzutsev, A., Trinchieri, G. ‘’Microbial DNA signature in plasma enables cancer diagnosis’’. Nat Rev Clin Oncol 17, 453–454 (2020). https://doi.org/10.1038/s41571-020-0391-1
- Nejman, D. et al ‘’The human tumor microbiome is composed of tumor type–specific intracellular bacteria’’. Science, 368:6494, 29 May 2020, p. 973-980, DOI: 10.1126/science.aay9189
- Hermida, L. et al ‘’Analyzing the tumor microbiome to predict cancer patient survival and drug response’’. Biorxiv, 25 July 2020. doi: https://doi.org/10.1101/2020.07.21.214148
- Duong, M.TQ., Qin, Y., You, SH. et al. ‘’Bacteria-cancer interactions: bacteria-based cancer therapy’’. Exp Mol Med 51, 1–15 (2019). https://doi.org/10.1038/s12276-019-0297-0
- Riquelme. E. et al. ‘’Tumor Microbiome Diversity and Composition Influence Pancreatic Cancer Outcomes’’. Cell. 2019 Aug 8;178(4):795-806.e12. doi: 10.1016/j.cell.2019.07.008. PMID: 31398337; PMCID: PMC7288240.
- Peters. BA et al. ‘’The Microbiome in Lung Cancer Tissue and Recurrence-Free Survival’’. Cancer Epidemiol Biomarkers Prev. 2019 Apr;28(4):731-740. doi: 10.1158/1055-9965.EPI-18-0966. Epub 2019 Feb 7. PMID: 30733306; PMCID: PMC6449216.
- Zhang, Χ.et al. “Pancreatic Cancer, Gut Microbiota, and Therapeutic Efficacy.” Journal of Cancer vol. 11,10 2749-2758. 20 Feb. 2020, doi:10.7150/jca.37445
- Mager, L. et al ‘’Microbiome-derived inosine modulates response to checkpoint inhibitor immunotherapy’’ Science, 369:6510, 18 September 2020, p. 1481-1489, DOI: 10.1126/science.abc3421
- Anonymous ‘’Takeda and Debiopharm agree licencing deal for GI targeted novel microbiome therapeutics’’ Microbiome Post, 16 June 2020, https://microbiomepost.com/takeda-and-debiopharm-agree-licencing-deal-for-gi-targeted-novel-microbiome-therapeutics/
- Ibid
- Anonymous ‘’BiomX Enters Collaboration with Boehringer Ingelheim with the Goal of Discovering Microbiome-Based Biomarkers for Inflammatory Bowel Disease’’. Businesswire, 2 September 2020, https://www.businesswire.com/news/home/20200902005061/en/BiomX-Enters-Collaboration-Boehringer-Ingelheim-Goal-Discovering).
- Fraser C. et al. ‘’Evaluation of a card collection-based faecal immunochemical test in screening for colorectal cancer using a two-tier reflex approach’’. Gut. 2007 Oct;56(10):1415-8. doi: 10.1136/gut.2007.119651. Epub 2007 Feb 19. PMID: 17309886; PMCID: PMC2000260.
- Chénard, T., Malick, M., Dubé, J. et al. ‘’The influence of blood on the human gut microbiome’’. BMC Microbiol 20, 44 (2020). https://doi.org/10.1186/s12866-020-01724-8
- Gosálbez, L., ‘’ The Microbiome Drug Landscape report: Promising clinical performance and signs of a maturing industry’’, The Microbiome Times, 26 October 2020, https://www.microbiometimes.com/the-microbiome-drug-landscape-report-promising-clinical-performance-and-signs-of-a-maturing-industry-4/
- Second Genome ‘’Gilead Sciences and Second Genome Announce Strategic Collaboration in Biomarker and Inflammatory Bowel Disease Drug Discovery (Press Release)’’ 6 April 2020, https://www.secondgenome.com/news/gilead-sciences-and-second-genome-announce-strategic-collaboration-in-biomarker-and-inflammatory-bowel-disease-drug-discovery
- Ibid 84
- Anonymous ‘’Persephone Biosciences Initiates the ARGONAUT Study of Gut Microbiome-Linked Immune Modulation in Cancer Treatment Response, 23 November 2020, https://www.microbiometimes.com/persephone-biosciences-initiates-the-argonaut-study-of-gut-microbiome-linked-immune-modulation-in-cancer-treatment-response/
- Tan, T. ‘’Bio-Me, Siolta partner on paediatric allergy and asthma risk test’’. First Word Medtech,
- Microbiome Drug Database, The Microbiome Times, https://www.microbiometimes.com/microbiome-drug-database/
- Manor, O., Dai, C.L., Kornilov, S.A. et al. ‘’Health and disease markers correlate with gut microbiome composition across thousands of people’’. Nat Commun 11, 5206 (2020). https://doi.org/10.1038/s41467-020-18871-1

Luis Gosálbez
Sandwalk BioVentures is a specialty strategy, innovation, regulatory and management consulting firm focused on microbiome technologies servicing companies in the food and pharma sectors, as well as financial and strategic investors exploring to enter this field. The company has created the Microbiome Drug Database™, an online repository containing the most extensive and thorough analysis of biotechnology companies developing pharmaceuticals from or through the microbiome.