
The advent of metagenomic sequencing has led to significant advancements in our understanding of the microbiome in a wide variety of contexts, from the human body1 to farm animals2,3, soil, and marine environments4. Each unique environment provides its own set of challenges in accurate microbial identification, including low microbial biomass in ocean samples, PCR inhibitors in soil samples, and high host DNA in certain human and animal samples. Sequencing of the 16S rRNA gene has been a popular and low-cost way to identify microbes in these samples. However, this method is limited by PCR primer and amplification bias along with unreliable identification below the genus level5. 16S rRNA gene sequencing studies that use primer sets targeting different variable regions cannot be compared directly as different regions selectively detect different bacterial taxa6. Further, relative abundance measures are inaccurate due to variation in the number of 16S rRNA gene operons present within differing bacterial species7.
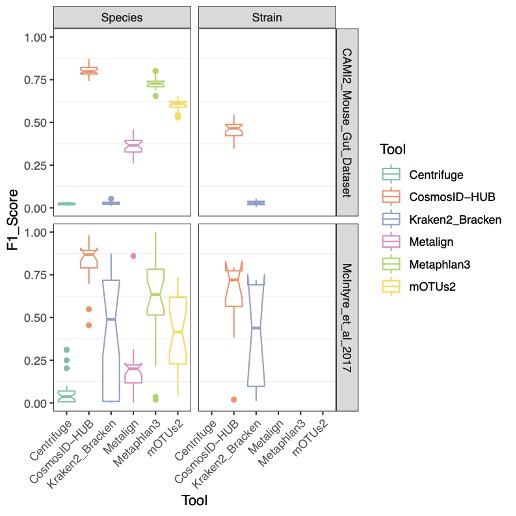
Whole genome shotgun sequencing (WGS) addresses the amplification, primer bias, and relative quantification issues by avoiding amplification altogether and sequencing the entire bacterial genome. Here, unique, single-copy marker gene sequences or reference genome sequences can be used to identify and quantify bacteria present within a sample more accurately. Because WGS relies on marker genes or alignment to a reference, it is possible to accurately identify microorganisms at the species or even strain taxonomic levels. However, there is wide variation in the accuracy of computational tools developed to perform these microbial identifications. Challenges faced in WGS methods include the requirement to account for variations in genetic diversity within species (i.e. some species are very diverse, whereas others are genetically uniform), mobile elements that are shared among species, the quality of reference genomes used, and the divergence of strains found in nature from the reference genomes that are used for identification. Here, we perform a benchmarking study to evaluate the performance of CosmosID-HUB to five other publicly available taxonomic classification algorithms Centrifuge11, Metaphlan312, Kraken2_Bracken13, mOTUs229 and Metalign30. These publicly available taxonomic classification algorithms are known for its high accuracy and precision when compared to other publicly available methods based on previous benchmarking evaluations8-10.An ideal metagenomics classifier will properly identify a large number of microorganisms while displaying a small number of false positives at all taxonomic levels. For this study, we used publicly available benchmarking datasets from CAMI2(Mouse Gut Dataset)27 and McIntyre et al 2017 benchmarking paper28, which consisted of mock communities of known compositions, to perform these comparisons.
Figure 1: F1 Scores at the Species and Strain Levels.
Importance of Strain Level Resolution
As metagenomics is increasingly becoming a method of choice across multi-disciplinary applications, the importance of sub-species and strain level variation is becoming ever more apparent.14-23 For example, specific strains of Streptococcus mutans produce hemorrhagic damage in the murine brain and other tissues18, whereas other strains are risk factors for ulcerative colitis.17 Likewise, different strains of the protozoan parasite Toxoplasma gondii manifest diverse pathologies and elicit altered host responses19. Particular variants of Staphylococcus epidermidis15 and Staphylococcus aureus16 affect virulence and biofilm formation. Certain strains of Bifidobacterium longum, but not others, protect against pathogens like Escherichia coli, and still others elicit differential immunomodulatory properties.14 Similarly, strain-specific immunomodulatory effects are seen for Propionibacterium freudenreichii21 and for another probiotic agent, Lactobacillus casei, variants derived from different ecological niches vary in their ability to bind foodborne carcinogens.22 The importance of strain resolution is much more apparent when assigning attribution, as exemplified in outbreaks of nosocomial infections such as Legionella pneumophila23-24 and Klebsiella pneumoniae.25 These examples serve to underscore why sub-species and strain level identification is so crucial to our understanding of microbial symbiosis and dysbiosis, and thus demonstrate the power of CosmosID-HUB metagenomics in defining the microbiome composition at a finer taxonomic resolution – critical information needed in microbiome research, epidemiological studies, microbial forensics, and outbreak investigations.
Access the Full Whitepaper Contact Cosmos ID
Footnotes
- Human Microbiome Project Consortium. Structure, function and diversity of the healthy human microbiome. Nature. 2012;486(7402):207-14.
- Kauter A, Epping L, Semmler T, Antao E-M, Kannapin D, Stoeckle SD, Gehlen H, Lübke-Becker A, Günther S, Wieler LH, Walther B. The gut microbiome of horses: current research on equine enteral microbiota and future perspectives. Anim Microbiome. 2019;1(1):14.
- Bergamaschi M, Tiezzi F, Howard J, Huang YJ, Gray KA, Schillebeeckx C, McNulty NP, Maltecca C. Gut microbiome composition differences among breeds impact feed efficiency in swine. Microbiome. 2020;8(1):110.
- Logares R, Deutschmann IM, Junger PC, Giner CR, Krabberød AK, Schmidt TSB, Rubinat-Ripoll L, Mestre M, Salazar G, Ruiz-González C, Sebastián M, de Vargas C, Acinas SG, Duarte CM, Gasol JM, Massana R. Disentangling the mechanisms shaping the surface ocean microbiota. Microbiome. 2020;8(1):55.
- Johnson JS, Spakowicz DJ, Hong B-Y, Petersen LM, Demkowicz P, Chen L, Leopold SR, Hanson BM, Agresta HO, Gerstein M, Sodergren E, Weinstock GM. Evaluation of 16S rRNA gene sequencing for species and strain-level microbiome analysis. Nat Commun. 2019;10(1):5029.
- Chakravorty S, Helb D, Burday M, Connell N, Alland D. A detailed analysis of 16S ribosomal RNA gene segments for the diagnosis of pathogenic bacteria. J Microbiol Methods. 2007;69(2):330-9.
- Acinas SG, Marcelino LA, Klepac-Ceraj V, Polz MF. Divergence and redundancy of 16S rRNA sequences in genomes with multiple rrn operons. J Bacteriol. 2004;186(9):2629-35.
- Sczyrba A, Hofmann P, Belmann P, Koslicki D, Janssen S, Dröge J, Gregor I, Majda S, Fiedler J, Dahms E, Bremges A, Fritz A, Garrido-Oter R, Jørgensen TS, Shapiro N, Blood PD, Gurevich A, Bai Y, Turaev D, DeMaere MZ, Chikhi R, Nagarajan N, Quince C, Meyer F, Balvočiūtė M, Hansen LH, Sørensen SJ, Chia BKH, Denis B, Froula JL, Wang Z, Egan R, Don Kang D, Cook JJ, Deltel C, Beckstette M, Lemaitre C, Peterlongo P, Rizk G, Lavenier D, Wu Y-W, Singer SW, Jain C, Strous M, Klingenberg H, Meinicke P, Barton MD, Lingner T, Lin H-H, Liao Y-C, Silva GGZ, Cuevas DA, Edwards RA, Saha S, Piro VC, Renard BY, Pop M, Klenk H-P, Göker M, Kyrpides NC, Woyke T, Vorholt JA, Schulze-Lefert P, Rubin EM, Darling AE, Rattei T, McHardy AC. Critical Assessment of Metagenome Interpretation-a benchmark of metagenomics software. Nat Methods. 2017;14(11):1063-71.
- Ye SH, Siddle KJ, Park DJ, Sabeti PC. Benchmarking metagenomics tools for taxonomic classification. Cell. 2019;178(4):779-94.
- Meyer F, Bremges A, Belmann P, Janssen S, McHardy AC, Koslicki D. Assessing taxonomic metagenome profilers with OPAL. Genome Biol. 2019;20(1):51.
- Kim D, Song L, Breitwieser FP, Salzberg SL. Centrifuge: rapid and sensitive classification of metagenomic sequences. Genome Res. 2016;26(12):1721-9.
- Beghini F, McIver LJ, Blanco-Míguez A, Dubois L, Asnicar F, Maharjan S, Mailyan A, Manghi P, Scholz M, Thomas AM, Valles-Colomer M, Weingart G, Zhang Y, Zolfo M, Huttenhower C, Franzosa EA, Segata N. Integrating taxonomic, functional, and strain-level profiling of diverse microbial communities with bioBakery 3. eLife. 2021;10.
- Wood DE, Lu J, Langmead B. Improved metagenomic analysis with Kraken 2. Genome Biol. 2019;20(1):257.
- Fukuda S, Toh H, Hase K, Oshima K, Nakanishi Y, Yoshimura K, Tobe T, Clarke JM, Topping DL, Suzuki T, Taylor TD, Itoh K, Kikuchi J, Morita H, Hattori M, Ohno H. Bifidobacteria can protect from enteropathogenic infection through production of acetate. Nature. 2011;469(7331):543-7.
- Gill SR, Fouts DE, Archer GL, Mongodin EF, Deboy RT, Ravel J, Paulsen IT, Kolonay JF, Brinkac L, Beanan M, Dodson RJ, Daugherty SC, Madupu R, Angiuoli SV, Durkin AS, Haft DH, Vamathevan J, Khouri H, Utterback T, Lee C, Dimitrov G, Jiang L, Qin H, Weidman J, Tran K, Kang K, Hance IR, Nelson KE, Fraser CM. Insights on evolution of virulence and resistance from the complete genome analysis of an early methicillin-resistant Staphylococcus aureus strain and a biofilm-producing methicillin-resistant Staphylococcus epidermidis strain. J Bacteriol. 2005;187(7):2426-38.
- Iwase T, Uehara Y, Shinji H, Tajima A, Seo H, Takada K, Agata T, Mizunoe Y. Staphylococcus epidermidis Esp inhibits Staphylococcus aureus biofilm formation and nasal colonization. Nature. 2010;465(7296):346-9.
- Kojima A, Nakano K, Wada K, Takahashi H, Katayama K, Yoneda M, Higurashi T, Nomura R, Hokamura K, Muranaka Y, Matsuhashi N, Umemura K, Kamisaki Y, Nakajima A, Ooshima T. Infection of specific strains of Streptococcus mutans, oral bacteria, confers a risk of ulcerative colitis. Sci Rep. 2012;2:332.
- Nakano K, Hokamura K, Taniguchi N, Wada K, Kudo C, Nomura R, Kojima A, Naka S, Muranaka Y, Thura M, Nakajima A, Masuda K, Nakagawa I, Speziale P, Shimada N, Amano A, Kamisaki Y, Tanaka T, Umemura K, Ooshima T. The collagen-binding protein of Streptococcus mutans is involved in haemorrhagic stroke. Nat Commun. 2011;2:485.
- Saeij JPJ, Boyle JP, Boothroyd JC. Differences among the three major strains of Toxoplasma gondii and their specific interactions with the infected host. Trends Parasitol. 2005;21(10):476-81.
- Sharon I, Morowitz MJ, Thomas BC, Costello EK, Relman DA, Banfield JF. Time series community genomics analysis reveals rapid shifts in bacterial species, strains, and phage during infant gut colonization. Genome Res. 2013;23(1):111-20.
- Foligné B, Deutsch S-M, Breton J, Cousin FJ, Dewulf J, Samson M, Pot B, Jan G. Promising immunomodulatory effects of selected strains of dairy propionibacteria as evidenced in vitro and in vivo. Appl Environ Microbiol. 2010;76(24):8259-64.
- Hernandez-Mendoza A, Garcia HS, Steele JL. Screening of Lactobacillus casei strains for their ability to bind aflatoxin B1. Food Chem Toxicol. 2009;47(6):1064-8.
- Helbig JH, Kurtz JB, Pastoris MC, Pelaz C, Lück PC. Antigenic lipopolysaccharide components of Legionella pneumophila recognized by monoclonal antibodies: possibilities and limitations for division of the species into serogroups. J Clin Microbiol. 1997;35(11):2841-5.
- Visca P, Goldoni P, Lück PC, Helbig JH, Cattani L, Giltri G, Bramati S, Castellani Pastoris M. Multiple types of Legionella pneumophila serogroup 6 in a hospital heated-water system associated with sporadic infections. J Clin Microbiol. 1999;37(7):2189-96.
- Snitkin ES, Zelazny AM, Thomas PJ, Stock F, NISC Comparative Sequencing Program Group, Henderson DK, Palmore TN, Segre JA. Tracking a hospital outbreak of carbapenem-resistant Klebsiella pneumoniae with whole-genome sequencing. Sci Transl Med. 2012;4(148):148ra116.

