
The overlap between the gut microbiome and tumor progression, metastasis and treatment sensitivity is one of the fastest growing areas of cancer research1. Recent studies have demonstrated that the microbiome plays important roles in oncogenesis processes, especially in the context of the so-called hallmarks of cancer. It also regulates the inpatient response to immune checkpoint inhibitors (ICIs), e.g. PD-1 inhibitors and CTLA-4 blockade, or even treatment-associated toxicity2-6. These findings have recognized microbiome signatures as a promising solution for the challenges of cancer drug development7.
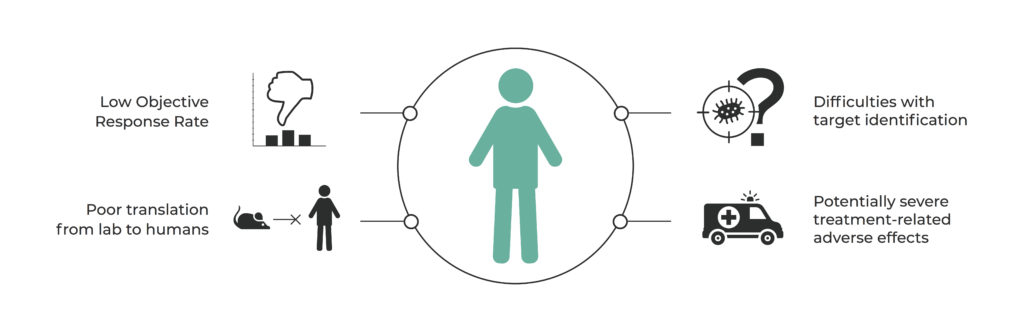
“How to modulate the human gut microbiome to improve response rates in cancer therapy?” currently seems to be the question that is driving research in a number of laboratories all over the world. The most advanced teams led by Jennifer Wargo, Laurence Zitvogel and Thomas Gajewski are exploring various paths to answer this question. This challenge consists of many aspects that can be addressed by microbiome signatures, including low objective response rates in phase II of clinical trials, poor translation from animal models to human studies, immunologically unresponsive ‘cold’ tumors, or treatment-related adverse effects such as colitis (Figure 1)8-11.
While all the aforementioned groups discovered a similar association between increased response rate and a beneficial gut microbiome composition, the definition of ‘beneficial’ in immuno-oncology remains unclear.
The published results define specific bacteria that distinguish immune checkpoint therapy responders, including Faecalibacterium prausnitzii, Gemmiger formicilis (Chaput et al.)12, Bacteroides thetaiotaomicron, Dorea formicogenerans (Frankel et al.)13, Bifidobacterium longum, Collinsella aerofaciens, Enterococcus faecium (Matson et al., 2018)3, and Akkermansia muciniphila (Routy et al.)14. Nevertheless, the specific bacterial taxa identified across different studies were not the same and so it was difficult to draw conclusions in relation to strains associated with responder groups5. This suggests that microbiota ‘signatures’ are a result not of taxonomy but of functional features (such as enhanced systemic immunity or intratumoral immune infiltration).
The limit of research in the taxonomic approach has been reached; this has resulted in researchers adopting slightly different ways of explaining the association between bacteria and cancer treatment. In the group led by Jennifer Wargo at MD Anderson (Houston, TX), the key focus was on the role of dietary factors and associated metabolic processes that lead to the production of effector agents by gut bacteria. On the other hand, researchers led by Thomas Gajewski at the University of Chicago (Chicago, IL) are investigating the role of non-bacterial members of the microbial community, namely viruses and phages. Another hypothesis drives the research of Laurence Zitvogel’s group at Gustave Roussy in Paris, which relies on analysis of the immunostimulatory agents produced by gut bacteria that may directly activate the immune system by mimicking tumor antigens.
As such a variety of potential links between microbiota and the immune system’s activity in cancer treatment have been observed, it is also worth utilizing modern computational approaches such as Big Data and Artificial Intelligence. Such unconstrained application of metagenomic sequencing data is not limited by any a priori developed hypothesis, therefore it may lead to the discovery of novel traits that will explain the ‘beneficial microbiome’.
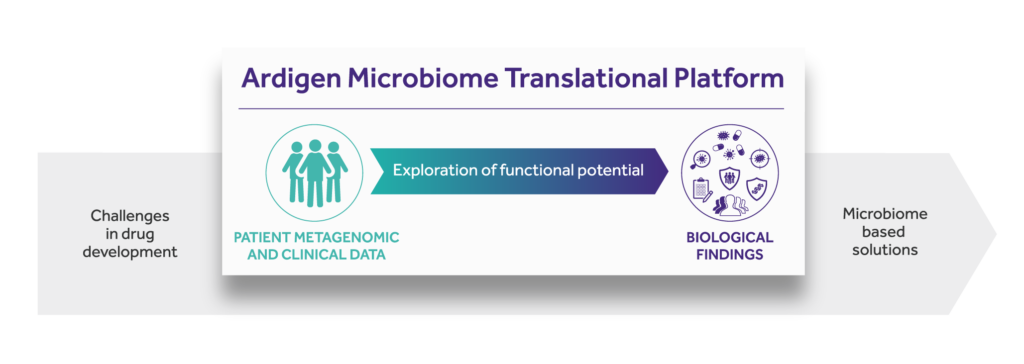
One of the first implementations of such an approach is Ardigen’s Microbiome Translational Platform. Ardigen addresses the aforementioned drug development challenges by starting with patients’ microbial and clinical data. We provide insight into the therapy-modulating interplay between host and microbiome by exploring the functional potential of commensal microbiota. In the process of revealing microbial signatures, we decode biological findings to clinical benefit.
Scientists have provided strong evidence that the microbiome has a mechanistic impact on anticancer processes in the human body. This has opened a new segment of digital microbiome-based diagnostics and therapeutics, such as microbial safety or response biomarkers, Next Generation Biotherapeutics (therapeutic probiotics, microbiome-derived small molecules or peptides), Live Biotherapeutic Products (single strain or bacterial consortia) and bacterial cancer vaccines (bacterial antigens stimulating immune system against cancer cells) (Figure 2)15. Ardigen has already applied this approach to data obtained from metastatic melanoma patients treated with anti-PD-1 therapy. We discovered, among others, that upregulation of the arginine synthesis pathway may play a role in the treatment response. These findings are in agreement with previous studies which reported that arginine enhances T-cell antitumor activity16, 17. On one hand, our approach expands the search space by exploring the full potential of the gut microbiome. On the other hand, this approach allows the costs of the necessary biological validation to be reduced and shows the great potential that will be available in the future design of innovative diagnostic and therapeutic solutions.
References:
[1] Gopalakrishnan, Vancheswaran, Beth A. Helmink, Christine N. Spencer, Alexandre Reuben, and Jennifer A. Wargo. “The Influence of the Gut Microbiome on Cancer, Immunity, and Cancer Immunotherapy.”Cancer Cell33, no. 4 (2018): 570–80. https://doi.org/10.1016/j.ccell.2018.03.015.
[2] Helmink, Beth A., M. A. Wadud Khan, Amanda Hermann, Vancheswaran Gopalakrishnan, and Jennifer A. Wargo. “The Microbiome, Cancer, and Cancer Therapy.” Nature Medicine25, no. 3 (2019): 377–88. https://doi.org/10.1038/s41591-019-0377-7.
[3] Matson, Vyara, Jessica Fessler, Riyue Bao, Tara Chongsuwat, Yuanyuan Zha, Maria-Luisa Alegre, Jason J. Luke, and Thomas F. Gajewski. “The Commensal Microbiome Is Associated with Anti–PD-1 Efficacy in Metastatic Melanoma Patients.” Science359, no. 6371 (April 2018): 104–8. https://doi.org/10.1126/science.aao3290.
[4] Gopalakrishnan, V., C. N. Spencer, L. Nezi, A. Reuben, M. C. Andrews, T. V. Karpinets, P. A. Prieto, D. Vicente, K. Hoffman, S. C. Wei, A. P. Cogdill, L. Zhao, C. W. Hudgens, D. S. Hutchinson, T. Manzo, M. Petaccia De Macedo, T. Cotechini, T. Kumar, W. S. Chen, S. M. Reddy, R. Szczepaniak Sloane, J. Galloway-Pena, H. Jiang, P. L. Chen, E. J. Shpall, K. Rezvani, A. M. Alousi, R. F. Chemaly, S. Shelburne, L. M. Vence, P. C. Okhuysen, V. B. Jensen, A. G. Swennes, F. Mcallister, E. Marcelo Riquelme Sanchez, Y. Zhang, E. Le Chatelier, L. Zitvogel, N. Pons, J. L. Austin-Breneman, L. E. Haydu, E. M. Burton, J. M. Gardner, E. Sirmans, J. Hu, A. J. Lazar, T. Tsujikawa, A. Diab, H. Tawbi, I. C. Glitza, W. J. Hwu, S. P. Patel, S. E. Woodman, R. N. Amaria, M. A. Davies, J. E. Gershenwald, P. Hwu, J. E. Lee, J. Zhang, L. M. Coussens, Z. A. Cooper, P. A. Futreal, C. R. Daniel, N. J. Ajami, J. F. Petrosino, M. T. Tetzlaff, P. Sharma, J. P. Allison, R. R. Jenq, and J. A. Wargo. “Gut Microbiome Modulates Response to Anti–PD-1 Immunotherapy in Melanoma Patients.” Science359, no. 6371 (2017): 97-103. doi:10.1126/science.aan4236.
[5] Fessler, Jessica, Vyara Matson, and Thomas F. Gajewski. “Exploring the Emerging Role of the Microbiome in Cancer Immunotherapy.” Journal for ImmunoTherapy of Cancer7, no. 1 (2019). doi:10.1186/s40425-019-0574-4.
[6] Mcquade, Jennifer L., Carrie R. Daniel, Beth A. Helmink, and Jennifer A. Wargo. “Modulating the Microbiome to Improve Therapeutic Response in Cancer.” The Lancet Oncology20, no. 2 (2019). doi:10.1016/s1470-2045(18)30952-5.
[7] Zitvogel, Laurence, Yuting Ma, Didier Raoult, Guido Kroemer, and Thomas F. Gajewski. “The Microbiome in Cancer Immunotherapy: Diagnostic Tools and Therapeutic Strategies.” Science359, no. 6382 (2018): 1366-370. doi:10.1126/science.aar6918.
[8] Elinav, Eran, Wendy S. Garrett, Giorgio Trinchieri, and Jennifer Wargo. “The Cancer Microbiome.” Nature Reviews Cancer19, no. 7 (2019): 371-76. doi:10.1038/s41568-019-0155-3.
[9] Rosshart, Stephan P., Jasmin Herz, Brian G. Vassallo, Ashli Hunter, Morgan K. Wall, Jonathan H. Badger, John A. Mcculloch, Dimitrios G. Anastasakis, Aishe A. Sarshad, Irina Leonardi, Nicholas Collins, Joshua A. Blatter, Seong-Ji Han, Samira Tamoutounour, Svetlana Potapova, Mark B. Foster St. Claire, Wuxing Yuan, Shurjo K. Sen, Matthew S. Dreier, Benedikt Hild, Markus Hafner, David Wang, Iliyan D. Iliev, Yasmine Belkaid, Giorgio Trinchieri, and Barbara Rehermann. “Laboratory Mice Born to Wild Mice Have Natural Microbiota and Model Human Immune Responses.” Science365, no. 6452 (2019). doi:10.1126/science.aaw4361.
[10] Wang, Yinghong, Diana H. Wiesnoski, Beth A. Helmink, Vancheswaran Gopalakrishnan, Kati Choi, Hebert L. Dupont, Zhi-Dong Jiang, Hamzah Abu-Sbeih, Christopher A. Sanchez, Chia-Chi Chang, Edwin R. Parra, Alejandro Francisco-Cruz, Gottumukkala S. Raju, John R. Stroehlein, Matthew T. Campbell, Jianjun Gao, Sumit K. Subudhi, Dipen M. Maru, Jorge M. Blando, Alexander J. Lazar, James P. Allison, Padmanee Sharma, Michael T. Tetzlaff, Jennifer A. Wargo, and Robert R. Jenq. “Fecal Microbiota Transplantation for Refractory Immune Checkpoint Inhibitor-associated Colitis.” Nature Medicine24, no. 12 (2018): 1804-808. doi:10.1038/s41591-018-0238-9.
[11] Dubin, Krista, Margaret K. Callahan, Boyu Ren, Raya Khanin, Agnes Viale, Lilan Ling, Daniel No, Asia Gobourne, Eric Littmann, Curtis Huttenhower, Eric G. Pamer, and Jedd D. Wolchok. “Intestinal Microbiome Analyses Identify Melanoma Patients at Risk for Checkpoint-blockade-induced Colitis.” Nature Communications7, no. 1 (2016). doi:10.1038/ncomms10391.
[12] Chaput, N., P. Lepage, C. Coutzac, E. Soularue, K. Le Roux, C. Monot, L. Boselli, E. Routier, L. Cassard, M. Collins, T. Vaysse, L. Marthey, A. Eggermont, V. Asvatourian, E. Lanoy, C. Mateus, C. Robert, and F. Carbonnel. “Baseline Gut Microbiota Predicts Clinical Response and Colitis in Metastatic Melanoma Patients Treated with Ipilimumab.” Annals of Oncology28, no. 6 (2017): 1368-379. doi:10.1093/annonc/mdx108.
[13] Frankel, Arthur E., Laura A. Coughlin, Jiwoong Kim, Thomas W. Froehlich, Yang Xie, Eugene P. Frenkel, and Andrew Y. Koh. “Metagenomic Shotgun Sequencing and Unbiased Metabolomic Profiling Identify Specific Human Gut Microbiota and Metabolites Associated with Immune Checkpoint Therapy Efficacy in Melanoma Patients.” Neoplasia19, no. 10 (2017): 848-55. doi:10.1016/j.neo.2017.08.004.
[14] Routy, Bertrand, Emmanuelle Le Chatelier, Lisa Derosa, Connie P. M. Duong, Maryam Tidjani Alou, Romain Daillère, Aurélie Fluckiger, Meriem Messaoudene, Conrad Rauber, Maria P. Roberti, Marine Fidelle, Caroline Flament, Vichnou Poirier-Colame, Paule Opolon, Christophe Klein, Kristina Iribarren, Laura Mondragón, Nicolas Jacquelot, Bo Qu, Gladys Ferrere, Céline Clémenson, Laura Mezquita, Jordi Remon Masip, Charles Naltet, Solenn Brosseau, Coureche Kaderbhai, Corentin Richard, Hira Rizvi, Florence Levenez, Nathalie Galleron, Benoit Quinquis, Nicolas Pons, Bernhard Ryffel, Véronique Minard-Colin, Patrick Gonin, Jean-Charles Soria, Eric Deutsch, Yohann Loriot, François Ghiringhelli, Gérard Zalcman, François Goldwasser, Bernard Escudier, Matthew D. Hellmann, Alexander Eggermont, Didier Raoult, Laurence Albiges, Guido Kroemer, and Laurence Zitvogel. “Gut Microbiome Influences Efficacy of PD-1–based Immunotherapy against Epithelial Tumors.” Science 359, no. 6371 (2017): 91-97. doi:10.1126/science.aan3706.
[15] O’Toole, Paul W., Julian R. Marchesi, and Colin Hill. “Next-generation Probiotics: The Spectrum from Probiotics to Live Biotherapeutics.” Nature Microbiology 2, no. 5 (2017). doi:10.1038/nmicrobiol.2017.57.
[16] Fletcher, Matthew, Maria E. Ramirez, Rosa A. Sierra, Patrick Raber, Paul Thevenot, Amir A. Al-Khami, Dulfary Sanchez-Pino, Claudia Hernandez, Dorota D. Wyczechowska, Augusto C. Ochoa, and Paulo C. Rodriguez. “L-Arginine Depletion Blunts Antitumor T-cell Responses by Inducing Myeloid-Derived Suppressor Cells.” Cancer Research 75, no. 2 (2014): 275-83. doi:10.1158/0008-5472.can-14-1491.
[17] Geiger, Roger, Jan C. Rieckmann, Tobias Wolf, Camilla Basso, Yuehan Feng, Tobias Fuhrer, Maria Kogadeeva, Paola Picotti, Felix Meissner, Matthias Mann, Nicola Zamboni, Federica Sallusto, and Antonio Lanzavecchia. “L-Arginine Modulates T Cell Metabolism and Enhances Survival and Anti-tumor Activity.” Cell 167, no. 3 (2016). doi:10.1016/j.cell.2016.09.031.


